Development of a New 1,2,4-butanetriol Biosynthesis Pathway in an Engineered Homoserine-producing Strain of Escherichia coli
Author Information
Other Information
1
Hamburg University of Technology, Institute of Bioprocess and Biosystems Engineering, 21073 Hamburg, Germany
2
Present Address: Center of Synthetic Biology and Integrated Bioengineering, School of Engineering, Westlake University, Hangzhou 310024, China
*
Authors to whom correspondence should be addressed.
Received: 06 February 2023 Accepted: 05 May 2023 Published: 10 May 2023
© 2023 The authors. This is an open access article under the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/).
Synth. Biol. Eng.
2023,
1(1), 10007;
DOI: 10.35534/sbe.2023.10007
ABSTRACT:
1,2,4-butanetriol
(BT) is a compound of high interest with applications in pharmaceutical and
materials. In this work, we designed a novel biosynthetic pathway for BT from
glucose via a nonessential amino acid homoserine. This non-natural pathway used
an engineered phosphoserine transaminase (SerCR42W/R77W) to achieve
the deamination of homoserine to 4-hydroxy-2-oxobutanoic acid (HOBA). Three
consecutive enzymes including a lactate dehydrogenase, a 4-hydroxybutyrate
CoA-transferase and a bifunctional aldehyde/alcohol dehydrogenase are used to
catalyze HOBA to BT. To enhance the carbon flux to homoserine, a
homoserine-producing Escherichia coli was developed by improving the
overexpression of two relevant key genes metL and lysC (V339A).
The simultaneous overexpression of the genes encoding these enzymes for the
homoserine-derived BT pathway enabled production of 19.6 mg/L BT from glucose
in the homoserine-producing E. coli.
Keywords:
1,2,4-butanetriol; Synthetic pathway; Homoserine; E. coli
1. Introduction
1,2,4-Butanetriol (BT) is a valuable compound with many applications in pharmaceuticals and materials field [1,2]. Among the various applications, BT is best known as a precursor for the production of 1,2,4-butanetriol trinitrate (BTTN), which can be used as propellant and energetic plasticizer because of its excellent characteristics [2,3,4]. BT is traditionally manufactured by reducing esterified malic acid under NaBH4 under harsh conditions [5,6]. However, this process generates multiple tons of borate salts as byproducts for the production of each ton of BT, thus causing severe environmental pollution and high production costs [7].
Microbial synthesis of BT from D-xylose or L-arabinose was first reported by Niu et al. (2003) [2]. Thereafter, a series of genetic engineering strategies were employed to improve the production of BT from xylose in engineered *Escherichia coli* (*E. coli*) [7,8,9]. A patent by Frost and Niu (2011) claimed a BT titer of 18 g/L and a yield of 0.55 mol/mol xylose in an *E. coli* strain with *yia*E and *ycd*W knockouts [7]. Bamba et al. (2019) [10] engineered *Saccharomyces cerevisiae* to produce 1.7 g/L of BT from 10 g/L xylose with a molar yield of 24.5%, after the enhancement of uptaking ability of Fe to improve the activity of the rate-limiting enzyme, D-xylonate dehydratase (XylD), and screen the optimal 2-ketoacid decarboxylase. Besides this, Wang et al. (2022) [11] constructed a synthetic pathway for the biosynthesis of BT from D-arabinose in *E. coli*, achieving 2.24 g/L BT after 48 h of catalysis under the optimized fermentation conditions.
Except for the bacterial synthesis of BT from xylose or arabinose, Abdel-Ghany et al. (2013) [3] introduced genes from synthetic xylose pathways into Arabidopsis plants for BT production. As a proof-of-concept, the engineered plants can synthesize 20 µg of BT per gram of soil-grown plants. Efforts have also been made to directly produce BT from glucose in *E. coli* strain. Li et al. (2014) [12] designed a new artificial pathway for the biosynthesis of BT from malate with 120 ng/L BT production. In this pathway, the low efficiency of CoA-dependent reaction of the first step, malyl CoA formation from malate catalyzed by malate thiokinase, was considered to be the bottleneck of the entire pathway.
Homoserine, a C4 intermediate for the biosynthesis of threonine and methionine in *E. coli*, has been used as a precursor for producing 1,3-propanediol (1,3-PDO), acrylate, 3-hydroxypropionate and 2,4-dihydroxybutyrate (DHB) in genetically modified microorganisms [13,14,15,16,17]. The biosynthesis pathway of homoserine consists of four enzymatic steps from oxaloacetate (OAA) and is strictly regulated by methionine, threonine, and lysine [18,19,20,21,22]. Aspartokinase (AK) isozymes are the key enzymes involved in this pathway. AK I (homoserine dehydrogenase I, ThrA, encoded by *thr*A) and AK II (homoserine dehydrogenase II, MetL, encoded by *met*L) are bifunctional enzymes (Figure 1). Both of them catalyze the second step and the final step in the biosynthesis of homoserine from OAA. AK III, which only catalyzes the second step, is encoded by *lys*C and sensitive to lysine. In *E. coli*, the expression of MetL is repressed by methionine and the activity of ThrA is strongly feedback-inhibited by threonine [23,34].
Recently, the substrate specificity of a phosphoserine aminotransferase (SerC) from *E. coli* was successfully altered from phosphoserine to homoserine. The engineered SerC can dramatically improve the production of 1,3-PDO in a homoserine-derived pathway in *E. coli* [17], making homoserine an appealing precursor for the biosynthesis of BT. The non-natural 4-hydroxy-2-oxobutanoic acid (HOBA), which is from the deamination of homoserine, differs from BT by only one terminal carboxyl group and one carbonyl group. Hence the conversion of HOBA to BT requires reduction of these two functional groups to hydroxy groups. The carbonyl group of HOBA can be reduced to a hydroxy group by lactate dehydrogenase A (LdhA) from *Lactococcus lactis* by structure-based enzyme engineering [15]. Studies carried out by Walther et al. (2018) [15] revealed that LdhAQ85C accepted HOBA and the natural substrate pyruvate with almost equal specificity, thus supporting an efficient conversion of HOBA to 2,4-dihydroxybutarate (DHB). The carboxyl group of DHB to be reduced to a hydroxy group needs three enzymatic reactions, which are sequentially catalyzed by 4-hydroxybutyrate CoA-transferase AbfT-2 from *Porphyromonas gingivalis* and the bifunctional aldehyde/alcohol dehydrogenase AdhE2 from *Clostridium acetobutylicum*. The non-natural DHB serves as a key intermediate in the biosynthetic BT pathway via malate. Studies showed that the recombinant *E. coli* expressing AbfT2 and AdhE2 could achieve 55 mg/L BT production after 24 h post-induction cultivation with the addition of 100 mM DHB, confirming the functionality of pathway from DHB to BT catalyzed by AbfT2 and AdhE2 [12]. Herein, BT production from glucose via extension of the homoserine pathway is enabled by the introduction of a five-step pathway composed of SerCR42W/R77W, LdhAQ85C, AbfT2 and AdhE2.In this work, a new biosynthesis pathway for BT production from glucose via homoserine was proposed and verified in *E. coli* (Figure 1). The key module of the BT biosynthesis pathway involves five consecutive enzymatic reactions. To test this novel BT pathway, a strain capable of significantly accumulating intracellular homoserine was constructed.
Figure 1. A new biosynthetic pathway for BT production from glucose via homoserine. Nonnative synthetic reactions are shown in red, natural homoserine pathway reactions are shown in green and other natural reactions are shown in black. Overexpressed enzymes for the homoserine pathway are shown in bold black. Red cross means blockage of the corresponding reaction. Major genes and the enzymes encoded: *ser*C: phosphoserine aminotransferase; *ldh*A: lactate dehydrogenase; *abfT-2*: 4-hydroxybutyrate CoA-transferase; *adhE2*: aldehyde/alcohol dehydrogenase; *lys*C: Aspartate kinase III; *met*L: Aspartate kinase II/homoserine dehydrogenase II.
2. Materials and Methods
*2.1. Strains and Plasmids Construction*
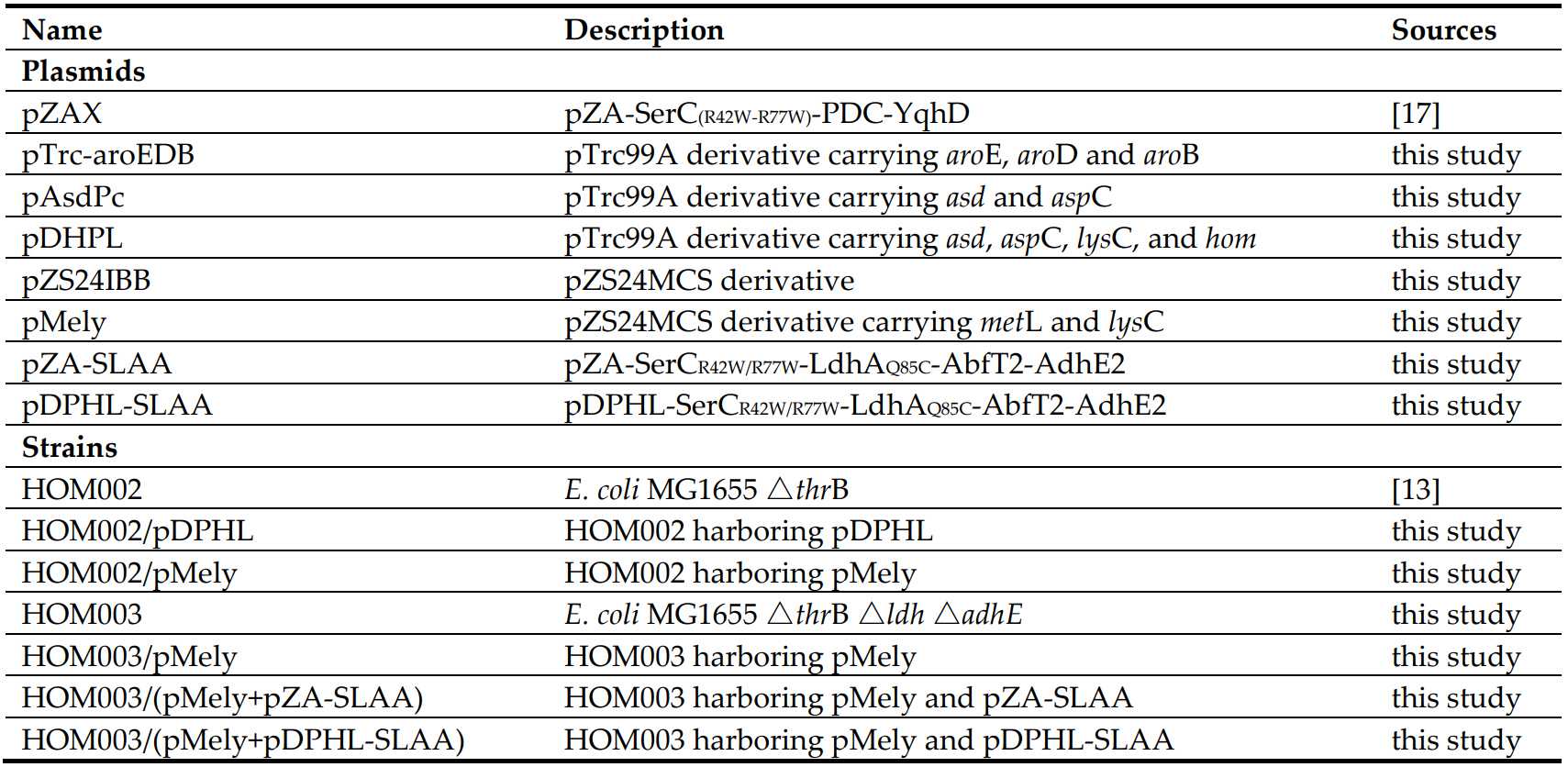
Strains and plasmids that were constructed and used in this study are listed in Table 1. The wild-type genes *asd* and *asp*C were amplified from *E. coli* MG1655 by PCR and inserted into plasmid pTrc-aroEDB, generating plasmid pAsdPc. The genes *hom* (R441A) [25], *lys*C (V339A) [26] and *met*L, *lys*C (V339A) were obtained by PCR and inserted into the plasmids pAsdPc and pZS24IBB, respectively. As a result, the plasmids pDPHL and pMely were obtained. The plasmids pDPHL and pMely were integrated into the host *E. coli* HOM002 to generate the recombinant HOM002/pDPHL and HOM002/pMely respectively.The gene *ser*C(R42W-R77W) was amplified from the plasmid pZAX, *ldh*A(Q85C) was codon optimized and chemically synthesized, *abfT2* was obtained from *Porphyromonas gingivalis* and *AdhE2* was obtained from *C. acetobutylicum*. All four genes were sequentially assembled into the plasmids pDPHL and pZA, resulting in pDPHL-SerCR42W/R77W-LdhAQ85C-AbfT2-AdhE2 (referred as pDPHL-SLAA) and pZA-SerCR42W/R77W-LdhAQ85C-AbfT2-AdhE2 (referred as pZA-SLAA) respectively. *E. coli* MG1655 △*thr*B △*ldh* △*adh*E (referred as HOM003) was obtained by deleting the genes *ldh* and *adh*E from the genome of HOM002 using a CRISPR/Cas9 system [26]. The plasmids pZA-SLAA and pDPHL-SLAA were introduced into the host HOM003/pMely to generate the recombinant strains HOM003/(pMely + pZA-SLAA) and HOM003/(pMely + pDPHL-SLAA) respectively.
*2.2. Cultivation Conditions*
For batch fermentation in bioreactor, the seed culture and the fermentation medium were modified from literature [25,27]. 60 g/L initial glucose was used and 1 g/L threonine was added in the fermentation medium. The seed culture was inoculated into a bioreactor filled with 2 L of fermentation medium with an initial OD600 of 0.1. The pH was adjusted to 7.4 with 25% NH4OH and 3 M of H3PO4 during fermentation. When the OD600 reached about 0.6, 0.2 mM IPTG was added.
The production of BT was performed in a modified M9 minimal medium containing 20 g/L glucose, 6.78 g/L Na2HPO4, 3.0 g/L KH2PO4, 0.5 g/L NaCl, 1.0 g/L NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 10 mM NaHCO3, 10 µg/mL thiamine, 1 g/L L-threonine and 100 mM MOPS [12]. The MOPS was used to improve the buffering capacity. Microaerobic conditions were established by flushing capped anaerobic bottles with nitrogen for at least 5 min and then piercing the septum with a 23G needle (Becton-Dickenson). The needle allows a small amount of air to enter the bottle [28].
Fermentations performed in DASGIP bioreactors used a differently modified M9 minimal medium containing 6.78 g/L Na2HPO4, 3.0 g/L KH2PO4, 0.5 g/L NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 2.0 g/L NH4Cl, 1.0 g/L (NH4)2SO4, supplemented with 20 g/L glucose and 1 g/L L-threonine [28]. The pH was kept at 7.0 using 2 M NH4OH (aerobic phase) or Na2CO3 (microaerobic phase). When the OD600 reached 10 during aerobic phase, 0.2 mM IPTG was added in the culture. After 1 h post-induction, the airflow rate was set to 0.02 vvm for microaerobic conditions, and 10 mM NaHCO3 was added. The agitation rate was set at 700 rpm. Glucose concentration in the culture was kept between 0.5 and 5.0 g/L.
*2.3. Analytical Methods*
Cell growth was monitored by measuring the optical density at 600 nm (OD600) using a spectrophotometer. For determining the correlation between the cell dry weight (CDW) and OD600, 2 mL of sample were pipetted into a pre-dried EP tube in triplicates, then centrifuged at 13,000 rpm for 5 min, and the supernatant was discarded. The cells were washed with 9% NaCl once and collected by centrifugation, then dried at 80 °C for more than 24 h after carefully removing the wash solution. The weight of each tube containing the dried cells was measured with an analytical balance. The correlation coefficient of CDW was defined as gram per liter per OD600 (g/L/ OD600).
For the measurement of homoserine concentration in the culture broth, filtered samples were used for analysis in an Ultimate 3000 HPLC (Dionex/Thermo Fisher) with a FLD 3100 detector and a Kinetex® 2.6 µm C18 100 Å, LC Column of 100 × 4.6 mm, Ea (Phenomenex). The derivatizing reagent (AccQ-Fluor Reagent Kit) was from Waters. Eluents included Eluent A of 140 mM Na-Acetate, 0.1% ACN, pH 4.95, Eluent B of 40% Milli-Q H2O, 60% ACN, and Eluent C of 95% ACN. The running program was set as follows: flow rate 1 mL/min, column oven temperature kept at 45 °C, injection volume 10 µL, and a different gradient concentration of Eluent A, Eluent B and Eluent C over time.
The products DHB and BT in culture broth samples were derivatized bytrimethylsilylation and quantitatively analyzed with GC, using a modified procedure adapted from literature [28,29]. 100 µL filtered samples were dried down in a speedvac concentrator (Martin Christ, RVC 2-25 CD-plus) for approximately 1 h at ambient temperature. Then 20 µL 10 mM cyclohexanol solution (as an internal standard) in dimethylformamide was added. Further, 100 µL N,O-bis(trimethylsilyl)trifluoro-acetimide (BSTFA) with 1% trimethylchlorosilane, was added, and the mixture was incubated at 70 °C for 30 min. The derivatized samples were centrifuged at 13,000 rpm for 5 min and then injected into GC. A Rtx-5SIL MS capillary column (Restek, 30 m × 0.25 mm ID × 0.25 µm film thickness) was used.
*2.4. Fast Sampling and Analysis*
Samples for the measurement of the concentrations of intracellular metabolites were taken from the reactor (Bioengineering AG, working volume 2 L) with a rapid sampling unit (RSU). With this RSU, the automatic fast sampling and on-filter quenching can be finished within 6 seconds as reported previously by da Luz et al. (2014) [30]. The preparations of mixed solution and wash solution and the extraction of the intracellular metabolites were described in Chen (2016) [25].
The calculation of the intracellular concentration of a particular metabolite (*Ci*) was performed with the following equation:N (mmol) is the total absolute amount of the given metabolite in the total extracted cells and F (liter) is the estimated total intracellular volume of the extracted cells. F is calculated on the basis of the dry weight of the extracted cells multiplied by the ratio of their volume to cellular dry weight. For *E. coli*, the ratio of cell volume to cellular dry weight is taken as 0.0023 liter per gram [31]. The correlation relating the dry cell weight to the optical density at 600 nm (OD600) for the homoserine-producing strain HOM002/pDPHL was determined as 0.366 ± 0.008 g/L/OD600 [25].
*2.5. Thermodynamic Analysis of the BT Pathway*
According to the group contribution method, some thermodynamic properties of the structures within the compounds are similar in different molecules [32]. Thus, the molecular structure of a given compound can be decomposed into a set of smaller substructures. Then the contributions from each of the substructures (or groups) are summed up to estimate the standard Gibbs free energy of formation (∆f*G*'°) of the compound. Similarly, the standard Gibbs free energy of reaction (∆r*G*'°) is estimated by summing the contribution of each structural group created or destroyed during the reaction. The contribution values used here are from Jankowski et al. (2008) [32].
Table 1. Strains and plasmids used in this study.
*Ci* = N/F
3. Results and Discussion
*3.1. Growth and Glucose Consumption of Recombinant E. coli HOM002/pDPHL*
To investigate the extra- and intracellular homoserine accumulation of the strain HOM002/pDPHL, a batch fermentation with 60 g/L glucose was carried out in bioreactor. As shown in Figure 2, the strain kept growing for about 1 h after adding IPTG into the medium, and then stopped growing for 4–5 h to adapt the carbon flux redirections. Then the growth was recovered and turned into an exponential growth phase with a specific growth rate 0.173 h−1 from 10 to 15 h (Figure 3). The off-gas analysis showed that the production of CO2 dropped sharply at 17.4 h and then kept decreasing till the end of fermentation. However, the glucose consumption rate was not significantly reduced. The maximal OD600 achieved in this cultivation was 16.4 ± 0.23 at 36 h during the stationary phase. The decline phase started at about 40 h with a negative growth rate (Figure 3).*3.2. The Extra- and Intracellular Concentrations of Homoserine*
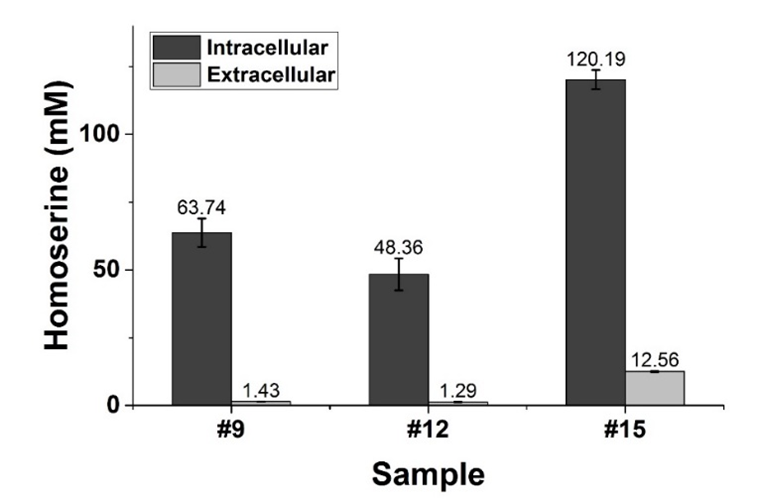
The strain HOM002/pDPHL cannot synthesize threonine due to the defective threonine biosynthetic pathway. Therefore, 1 g/L of threonine was added into the initial medium. From 13 to 15.5 h (#9 to #12 in Figure 2), the biomass (OD600) was increased from 5.03 ± 0.12 to 7.93 ± 0.03, whereas the intra- and extracellular concentration of threonine was dramatically decreased from 95.55 ± 7.83 to 37.11 ± 3.46 mM and 4.09 ± 0.09 to 2.14 ± 0.05 mM, respectively. After 17.4 h, the shortage of threonine became a factor in limiting the cell growth and caused the change of CO2. As for homoserine from 15.5 to 24 h, the extracellular homoserine was significantly increased (from 1.29 ± 0.06 to 12.56 ± 0.24 mM) by about 10 times. After that, it kept increasing almost till the end of the fermentation. Finally, about 2.8 g/L extracellular homoserine was obtained with a glucose conversion rate of 4.7% (g/g). And the overall productivity of extracellular homoserine was only about 0.037 g/L/h. It was also found that homoserine was quickly and significantly accumulated in the cell, even though the extracellular concentration of homoserine was very low before 15.5 h. As shown in Figure 4, the intracellular concentrations of homoserine are much higher than the corresponding extracellular ones. The ratio of intracellular to extracellular concentrations varied from 9.5 to 44.5, suggesting insufficiency of permease which is responsible for transferring homoserine out of the cell [33,34,35]. *3.3. Comparison of Two Constructed Homoserine-producing Strains*
Considering that the simultaneous overexpression of four proteins from the plasmid pDPHL may represent a huge metabolic burden for cells and limit the performance of heterologous BT pathway in *E. coli*, another plasmid pMely with only the genes of *lys*C and *met*L was constructed. Both AK III (LysC) and homoserine dehydrogenase (MetL) are crucial in the homoserine biosynthesis pathway. Enhancing the reactions catalyzed by LysC and MetL was speculated to efficiently improve the production of homoserine. To this end, comparison of HOM002/pDPHL and HOM002/pMely was carried out at different pH values in batch fermentations.
As illustrated in Figure 5, glucose was consumed faster for both strains at pH 7.4. At the same level of pH, HOM002/pMely performed better than HOM002/pDPHL, since it grew faster and reached much higher OD600. The expression of four genes which were all equipped with a strong rbs and controlled by a strong promoter *trc* or *tac* in the plasmid pDPHL may result in a substantial metabolic burden on cells and limit its growth. As a result, HOM002/pMely exhibited a higher production of homoserine, as shown in Figure 5, making it a more potent host strain for the biosynthesis of BT. In addition, the genes *ldh*A and *adh*E encoding lactate dehydrogenase and aldehyde/alcohol dehydrogenase, respectively, were removed from the *E. coli* genome to increase NADH supplementation and decrease byproducts formation under oxygen-limited conditions. The resulting host HOM002/pMely △*adh*E △*ldh*A was named as HOM003/pMely.*3.4. Construction of a Nonnative Biosynthetic BT Pathway from Homoserine in E. coli*
The thermodynamic feasibility of the proposed BT pathway was analyzed using the group contribution method [32]. The calculated total standard Gibbs free energy change of the five reactions for BT production from homoserine is −16.6 kcal/mol, suggesting that the designed BT pathway is thermodynamically feasible (Figure 6). However, the reactions of homoserine conversion to 4-hydroxy-2-oxobutanoate and 2,4-dihydroxybutyrl-CoA conversion to 2,4-dihydroxybutyraldehyde have zero to positive standard Gibbs free energy of reaction (∆r*G*'°), indicating that a push-and-pull strategy is necessary for these reactions to proceed in the desired direction and with adequate fluxes.Considering the necessity of the pMely plasmid in the host strain HOM002/pMely, to facilitate the construction and to keep plasmid stability during cultivation, the entire BT pathway was constructed in one plasmid. The pZA and pDPHL plasmids, which are compatible with pMely in *E. coli*, were used. The gene of LdhAQ85C was codon- optimized and the Gln85Cys point mutation was used, which showed an increased activity towards 4-hydroxy-2-oxobutanoate and a decreased activity towards the natural substrate pyruvate [35]. Finally, the resulting plasmids pZA-SLAA and pDPHL-SLAA were obtained (see Table 1).
*3.5. Production of BT from Glucose*
To verify the capability of the homoserine-derived pathway for BT production *in vivo*, two recombinant strains HOM003/(pMely+pZA-SLAA) and HOM003/(pMely+pDPHL-SLAA) were investigated using glucose as the sole carbon source respectively. The strain HOM003/pMely harboring no any BT biosynthetic pathway was used as the control strain. However, there was no detectable BT produced in batch fermentations with these strains, when they were cultivated in anaerobic bottles containing modified M9 medium. The enzymatic reactions catalyzed by AbfT2 and AdhE2 in the downstream pathway from DHB to BT, were supposed to be the rate-limiting steps in the BT pathway. The enzyme AbfT2 exhibited 4-hydroxybutyryl-CoA transferase activity when implemented as part of a pathway to produce 1,4-butanediol [28], but its activity towards the non-natural intermediate DHB is rarely reported. The enzyme AdhE2 from *C. acetobutylicum* is oxygen sensitive and contains a highly conserved iron-binding site. The anaerobic environment may severely affect the activity of AdhE2.
To better validate the feasibility of the BT biosynthetic pathway, we studied the recombinant strains in fed-batch fermentation (see Figure 7). Fermentations were performed with 1 L initial culture volume using modified M9 minimal medium supplemented with 20 g/L glucose. More specific operations referred to the fermentation are as described in Yim et al. (2011) [28]. According to the pathway proposed, three molecules of NADH and one molecule of ATP are required for the synthesis of one molecule of BT from homoserine. The supply of the reducing equivalents should be fulfilled by glycolysis and the oxidative TCA cycle. However, under completely anaerobic conditions, the carbon flux through the TCA cycle will be blocked [36]. Therefore, on considering the constraints of AdhE2 oxygen-sensitivity and oxygen requirement for a functional TCA cycle, microaerobic fermentation was used for the synthesis of BT.
DHB concentration in the cultures was detected to be 745 ± 10.8 mg/L and 625.1 ± 8.1 mg/L after 54 h fermentations for the strains HOM003/(pMely+pZA-SLAA) and HOM003/(pMely+pDPHL-SLAA), respectively (see Figure 8). As expected, there was no detectable BT production in the control strain HOM/pMely. The production of BT in strains HOM003/(pMely+pDPHL-SLAA) and HOM003/(pMely+pZA-SLAA) reached 19.6 ± 5.9 mg/L and 8.9 ± 0.4 mg/L in the fed-batch fermentations, respectively (See Figure 9).
The poor overexpression of LdhAQ85C in the cells and its low affinity towards HOBA may limit the production of DHB. Recently, a more efficient malate dehydrogenase from *E. coli* to pull down the pathway in DHB production was reported by Frazão et al. (2018) [37]. The employment of this engineered malate dehydrogenase instead of LdhAQ85C could result in higher DHB titer, which would help to increase the concentration of the key intermediate DHB and be beneficial to achieve a higher BT production. AdhE2 is an oxygen-sensitive and is the only enzyme reported so far to have the ability to catalyze 2,4-dihydroxybutyrl-CoA to 2,4-dihydroxybutyraldehyde, followed by its subsequent reduction to BT. AdhE2 was firstly used in the 1,4-butanediol biosynthesis pathway in *E. coli* [28] and then employed in the BT pathway by Li et al. (2014) [12] with a very poor performance.
There’re three enzymatic reactions consuming NADH in the BT pathway. Low NADH/NAD+ ratio may limit the production of BT. Considering the requirement of NADH in the BT pathway, the conditions of microaerobic fermentation should be optimized to ensure the availability of sufficient NADH to drive the BT pathway. An alternative choice is the introduction of heterologous formate dehydrogenase (*fdh*) to recycle NAD+ to NADH by catalyzing formate to CO2 and keep redox rebalance [38,39]. On the other hand, re-engineering of the relevant enzymes in the BT pathway and optimization of the conditions for fed-batch fermentations are alternative ways to achieve a higher BT production.
Figure 2. Glucose consumption, growth and CO2 production of the recombinant *E. coli* HOM002/pDPHL during a batch fermentation in bioreactor. The initial glucose concentration was 60 g/L. The samples taken for quantifying intracellular metabolites were marked with arrows (gray), their corresponding fermentation time is listed as follows: #9 (13 h), #12 (15.5 h), #15 (27.2 h).
Figure 3. The growth rate of *E. coli* HOM002/pDPHL cultivated in bioreactor.
Figure 4. Comparison of the intra- and extracellular concentrations of homoserine. Homoserine was produced in the strain of HOM002/pDPHL.
Figure 5. Comparison of HOM002/pDPHL and HOM002/pMely at different pH values. (A) concentration of glucose in culture medium; (B) OD600; (C) concentration of homoserine in culture medium. Error bars represent standard deviations of the measurements performed in triplicate.
Figure 6. Calculated values of the standard Gibbs free energy of formation (∆f*G*'°) of compound, the standard Gibbs free energy of each reaction (∆r*G*'°) and the overall reaction (∆overall reaction*G*'°) of the proposed metabolic pathway from homoserine to BT.
Figure 7. Fermentation profile of the strains HOM003/(pMely+pZA-SLAA) and HOM003/(pMely+pDPHL-SLAA) for BT production.
Figure 8. GC analysis of DHB in the samples from fed-batch fermentation. (A) recombinant strain HOM003/(pMely+pZA-SLAA); (B) recombinant strain HOM003/(pMely+pDPHL-SLAA); The retention time of DHB is 11.50 min; Peaks marked blue are for DHB standard, the red ones are from samples.
Figure 9. GC analysis of BT in the samples from fed-batch fermentations. (A) recombinant strain HOM003/pMely; (B) recombinant strain HOM003/(pMely+pDPHL-SLAA); (C) recombinant strain HOM003/(pMely+pZA-SLAA); The retention time of BT is 11.00 min; Peaks marked blue are for BT standard, the red ones are from samples.
4. Conclusions
In this work, recombinant *E. coli* strains HOM002/pDPHL and HOM002/pMely capable of producing homoserine from glucose with a high efficiency were engineered by enhancing the homoserine biosynthetic pathway from oxaloacetic acid (OAA). The results showed that HOM002/pMely is more capable of accumulating homoserine. By using this homoserine-producing strain, we explored the feasibility of a synthetic pathway enabling BT biosynthesis from glucose via homoserine under microaerobic conditions by flushing with air at a rate of 0.02 vvm. This pathway was evaluated to be thermodynamically feasible as the standard Gibbs free energy for the overall reaction was estimated to be -16.6 kcal/mol. Considering that the expression of four genes under a strong promoter in multiple copy may result in a substantial metabolic burden on the cells, both medium-copy number plasmid pZA and pDPHL with different promoters were tried. The resulting strains HOM003/(pMely+pDPHL-SLAA) and HOM003/(pMely+pZA-SLAA) were able to produce 19.6 ± 5.9 and 8.9 ± 0.4 mg/L, respectively. The result is much higher than that of BT pathway from malate reported in literature (120 ng/L). This study shows the validity of a novel BT biosynthetic pathway from glucose via homoserine.
Acknowledgments
Y.Z. thanks the Chinese Scholarship Council for a Ph.D. scholarship. The authors thank Dr. Jie Ren for providing us with the plasmid carrying LdhAQ85C, Dr. Wei Wang, Anna Gorte, and Dipl.-Ing. Andrea Simon from the Central Analytic laboratory of TUHH for their assistance during this work.
Author Contributions
Y.Z.: Conceptualization, Investigation, Visualization, Writing-Original Draft Preparation; L.C.: Conceptualization, Investigation, Visualization; A.T.: Investigation, data curation; A.Z.: Conceptualization, Supervision, Funding Acquisition, Writing-Reviewing and Editing.
Ethics Statement
Not applicable.
Informed Consent Statement
Not applicable.
Funding
This research was funded by a PhD scholarship from the Chinese Scholarship Council and supported by the Center for Synthetic Biology and Integrated Bioengineering at Westlake University and Westlake Education Foundation.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
References
1.
Bhoge SM, Kshirsagar P, Richhariya S, Singh K. Process for the preparation of fosamprenavir calcium. US Patent 9085592B2, 2012.
2.
Niu W, Molefe MN, Frost JW. Microbial Synthesis of the Energetic Material Precursor 1,2,4-Butanetriol. J. Am. Chem. Soc. 2003, 125, 12998–12999. [Google Scholar]
3.
Abdel-Ghany SE, Day I, Heuberger AL, Broeckling CD, Reddy AS. Metabolic engineering of Arabidopsis for butanetriol production using bacterial genes. Metab. Eng. 2013, 20, 109–120. [Google Scholar]
4.
Sun L, Yang F, Sun H, Zhu T, Li X, Li Y, et al. Synthetic pathway optimization for improved 1,2,4-butanetriol production. J. Ind. Microbiol. Biot. 2016, 43, 67–78. [Google Scholar]
5.
Bal’zhinimaev BS, Paukshtis EA, Suknev AP, Makolkin NV. Highly selective/enantioselective Pt-ReOx/C catalyst for hydrogenation of L-malic acid at mild conditions. J. Energy Chem. 2018, 27, 903–912. [Google Scholar]
6.
Ikai K, Mikami M, Furukawa Y, Urano T, Ohtaka S. Process for Preparing 1,2,4-butanetriol. US Patent 6949684B2, 2005.
7.
Frost JW, Niu W. Microbial Synthesis of d-1,2,4-butanetriol. US Patent 2011/0076730 A1, 2011.
8.
Lu X, He S, Zong H, Song J, Chen W, Zhuge B. Improved 1,2,4-butanetriol production from an engineered Escherichia coli by co-expression of different chaperone proteins. World J. Microb. Biot. 2016, 32, 149. [Google Scholar]
9.
Jing P, Cao X, Lu X, Zong H, Zhuge B. Modification of an engineered Escherichia coli by a combined strategy of deleting branch pathway, fine-tuning xylose isomerase expression, and substituting decarboxylase to improve 1,2,4-butanetriol production. J. Biosci. Bioeng. 2018, 126, 547–552. [Google Scholar]
10.
Bamba T, Yukawa T, Guirimand G, Inokuma K, Sasaki K, Hasunuma T, et al. Production of 1,2,4-butanetriol from xylose by Saccharomyces cerevisiae through Fe metabolic engineering. Metab. Eng. 2019, 56, 17–27. [Google Scholar]
11.
Wang J, Chen QY, Wang X, Chen KQ, Ouyang PK. The biosynthesis of D-1,2,4-butanetriol from d-arabinose with an engineered Escherichia coli. Front. Bioeng. Biotechnol. 2022, 10, 844517. [Google Scholar]
12.
Li X, Cai Z, Li Y, Zhang Y. Design and construction of a non-natural malate to 1,2,4-butanetriol pathway creates possibility to produce 1,2,4-butanetriol from glucose. Sci. Rep. 2014, 4, 5541. [Google Scholar]
13.
Chen Z, Geng F, Zeng AP. Protein design and engineering of a de novo pathway for microbial production of 1,3-propanediol from glucose. Biotechnol. J. 2015, 10, 284–289. [Google Scholar]
14.
Jun X, Charles WS, Phillip RG, Velasquez, JE. Microorganisms and Methods for Producing Acrylate and Other Products from Homoserine. WIPO Patent WO2013052717, 2013.
15.
Walther T, Calvayrac F, Malbert Y, Alkim C, Dressaire C, Cordier H, et al. Construction of a synthetic metabolic pathway for the production of 2,4-dihydroxybutyric acid from homoserine. Metab. Eng. 2018, 45, 237–245. [Google Scholar]
16.
Walther T, Cordier H, Dressaire C, Francois JM, Huet R. Method for the preparation of 2,4-dihydroxybutyrate. WIPO Patent WO/2014/009435, 2014.
17.
Zhang YJ, Ma CW, Dischert W, Soucaille P, Zeng AP. Engineering of Phosphoserine Aminotransferase Increases the Conversion of l-Homoserine to 4-Hydroxy-2-ketobutyrate in a Glycerol-Independent Pathway of 1,3-Propanediol Production from Glucose. Biotechnol. J. 2019, 14, e1900003. [Google Scholar]
18.
Liu P, Zhang, B, Yao ZH, Liu, ZQ, Zheng, YG. Multiplex design of the metabolic network for production of l-homoserine in Escherichia coli. Appl. Environ. Microb. 2020, 86, e01477-20. [Google Scholar]
19.
Vo TM, Park S. Metabolic engineering Escherichia coli W3110 for efficient production of homoserine from glucose. Metab. Eng. 2022, 73, 104–113. [Google Scholar]
20.
Liu M, Lou JL, Gu JL, Lyu XM, Wang FQ, Wei DZ. Increasing L-homoserine production in Escherichia coli by engineering the central metabolic pathways. J. Biotechnol. 2020, 314–315, 1–7. [Google Scholar]
21.
Liu P, Liu JS, Zhang B, Liu ZQ, Zheng, YG. Increasement of O-acetylhomoserine production in Escherichia coli by modification of glycerol-oxidative pathway coupled with optimization of fermentation. Biotechnol. Lett. 2021, 43, 105–117. [Google Scholar]
22.
Zakataeva NP, Aleshin VV, Tokmakova IL, Troshin PV, Livshits VA. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 1999, 452, 228–232. [Google Scholar]
23.
Chen Z, Rappert S, Sun J, Zeng AP. Integrating molecular dynamics and co-evolutionary analysis for reliable target prediction and deregulation of the allosteric inhibition of aspartokinase for amino acid production. J. Biotechnol. 2011, 154, 248–254. [Google Scholar]
24.
Chen Z, Rappert S, Zeng AP. Rational design of allosteric regulation of homoserine dehydrogenase by a nonnatural inhibitor L-lysine. ACS Synth. Biol. 2015, 4, 126–131. [Google Scholar]
25.
Chen L. Rational metabolic engineering and systematic analysis of Escherichia coli for L-tryptophan bioproduction. PhD Thesis. Technische Universität Hamburg: Hamburg, Germany, 2016.
26.
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar]
27.
Chen L, Zeng AP. Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration. Appl. Microbiol. Biotechnol. 2017, 101, 559–568. [Google Scholar]
28.
Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011, 7, 445–452. [Google Scholar]
29.
Elliott S, Burgess V. The presence of gamma-hydroxybutyric acid (GHB) and gammabutyrolactone (GBL) in alcoholic and non-alcoholic beverages. Forensic Sci. Int. 2005, 151, 289–292. [Google Scholar]
30.
da Luz JA, Hans E, Zeng AP. Automated fast filtration and on‐filter quenching improve the intracellular metabolite analysis of microorganisms. Eng. Life Sci. 2014, 14, 135–142. [Google Scholar]
31.
Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 2008, 3, 1299–1311. [Google Scholar]
32.
Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 2008, 95, 1487–1499. [Google Scholar]
33.
Li H, Wang BS, Zhu LH, Cheng S, Li YR, Zhang L, et al. Metabolic engineering of Escherichia coli W3110 for L-homoserine production. Process Biochem. 2016, 51, 1973–1983. [Google Scholar]
34.
Sumantran VN, Schweizer HP, Datta P. A novel membrane-associated threonine permease encoded by the tdcC gene of Escherichia coli. J. Bacteriol. 1990, 172, 4288–4294. [Google Scholar]
35.
Templeton BA, Savageau MA. Transport of Biosynthetic Intermediates: Homoserine and Threonine Uptake in Escherichia coli. J. Bacteriol. 1974, 117, 1002–1009. [Google Scholar]
36.
Partridge JD, Sanguinetti G, Dibden DP, Roberts RE, Poole RK, Green J. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 2007, 282, 11230–11237. [Google Scholar]
37.
Frazão CJR, Topham CM, Malbert Y, François JM, Walther T. Rational engineering of a malate dehydrogenase for microbial production of 2,4-dihydroxybutyric acid via homoserine pathway. Biochem. J. 2018, 475, 3887–3901. [Google Scholar]
38.
Ma C, Ou J, Xu N, Fierst JL, Yang S-T, Liu X. Rebalancing Redox to Improve Biobutanol Production by Clostridium tyrobutyricum. Bioengineering 2016, 3, 2. [Google Scholar]
39.
Molla GS, Wohlgemuth R, Liese A. One-pot enzymatic reaction sequence for the syntheses of d-glyceraldehyde 3-phosphate and l-glycerol 3-phosphate. J. Mol. Catal. B-Enzym. 2016, 124, 77–82. [Google Scholar]