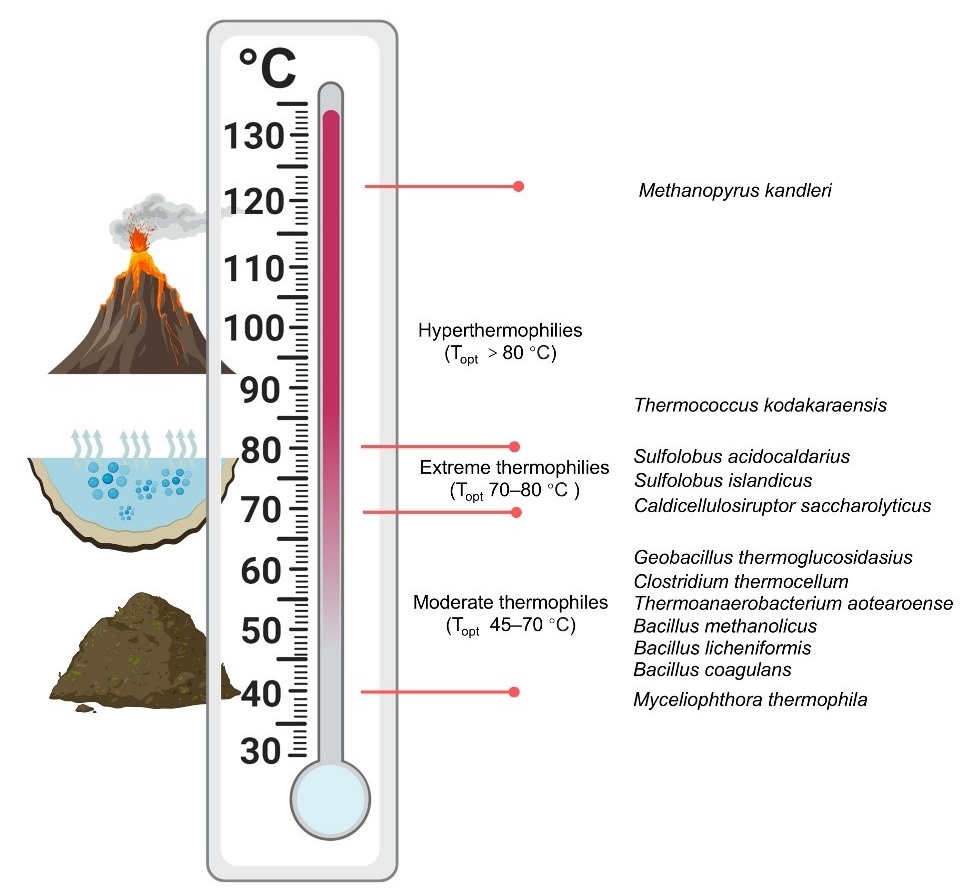
Thermophiles refer to a group of microorganisms with an optimal growth temperature (T
OPT) exceeding 45 °C [
14]. These organisms are widely distributed in high-temperature habitats on earth, including hot springs and hydrothermal vents, holding significant research value in fields such as biogeochemical cycles, microbial diversity, and microbial evolution [
15]. Additionally, high-temperature environments such as composting piles and industrial wastewater treatment systems also serve as habitats for thermophilic microorganisms (). Based on the growth temperature ranges, thermophiles are further classified into several groups. Microorganisms that thrive at temperatures between 45 °C and 70 °C are termed moderate thermophiles, while those that survive at temperatures from 70 °C to 80 °C are classified as extreme thermophiles. Organisms that flourish at temperatures above 80 °C are referred to as hyperthermophiles [
8,
16]. Notably, reports indicate that certain hyperthermophiles, such as
Methanopyrus kandleri, can survive at extreme temperatures ranging from 116 °C to 122 °C [
17].
. Growth temperature range and representative species of thermophilic microorganisms.
Thermophilic microorganisms demonstrate extensive potential for application across multiple industrial sectors, with a growing demand driven by technological advancements. For instance, certain thermophilic strains are capable of withstanding chemical denaturants and maintaining activity across a broad pH range, thereby enabling efficient production of biofuels such as ethanol and isobutanol under high-temperature conditions [
18]. Additionally, thermophiles are a significant source of a variety of thermally stable enzymes with high industrial value. These enzymes include but are not limited to, amylases, cellulases, xylanases, lipases, and DNA polymerases, whose stability in elevated temperatures provides unique advantages for industrial applications [
19].
2.1. Design Principles of High-Temperature Catalytic Platforms
To fully leverage the potential of thermophilic microorganisms and enzymes as catalysts, designing and optimizing high-temperature catalytic platforms requires consideration of catalyst types, reaction conditions, and catalyst combinations. Application scenarios are the primary consideration when selecting appropriate thermophilic catalysts. The primary reactions involved in high-temperature catalysis include hydrolysis, esterification, hydrogenation, and redox reactions, each suited to specific applications. For instance, hydrolysis breaks down complex polymers like starch, proteins, and cellulose, while esterification is central to lipid and biodiesel production [
11,
20,
21,
22]. Hydrogenation plays a role in hydrogen fermentation, and redox reactions are widely used in processes such as lignin degradation, wastewater treatment, metal bioleaching, cosmetic production, and methane oxidation [
23,
24,
25,
26,
27].Optimizing reaction conditions such as temperature, pH, reactant concentration, and catalyst concentration for different catalysts or catalytic reactions helps maximize their advantages and improve catalytic efficiency. Tailoring substrates based on microbial metabolic characteristics is also essential. For instance,
Clostridium thermocellum excels in lignin conversion, while
Bacillus licheniformis effectively metabolizes inulin and other non-sugar biomasses [
28,
29]. In specialized catalytic platforms, factors like nutrient limitation and light intensity enhance lipid accumulation in thermophilic algae, while hydrogenation reactions require careful consideration of reactor design, operational conditions, and safety precautions [
30,
31,
32]. Directly using thermophilic enzymes can be quite expensive due to their high cost, but the immobilized enzyme strategy helps mitigate this issue, as it is suitable for continuous production [
33]. For applications involving cascade reactions, combining microorganisms and thermozymes is an efficient strategy to create highly effective biocatalytic platforms. For example, expressing highly active thermophilic enzymes in non-thermophilic bacteria to develop high-temperature whole-cell catalytic platforms reduces side reactions, streamlines metabolism, and improves the selectivity of thermozymes catalysis [
34].
2.2. Thermophilic Microorganisms Serving as NGIB Platform
In the fermentation industry, high-temperature catalytic systems demonstrate significant energy-saving advantages, particularly in reducing cooling energy requirements and the risk of contamination, which is of great importance for industrial energy conservation and emission reduction [
35]. In recent years, one of the key research focuses in biotechnology has been the isolation and metabolic engineering of thermophilic microorganisms, which exhibit optimal growth performance at moderate to high temperatures ranging from 50 °C to 80 °C (). The application of thermophilic microorganisms as high-temperature catalysis platforms is shown in .
. Industrial applications of thermophilic microorganisms.
, as an efficient and safe high-temperature fermentation platform strain, grows rapidly at 50 °C. Its strong resistance to chemical denaturants and broad pH adaptability enable the effective production of biofuels, such as D-lactic acid and 2,3-butanediol, in high-temperature environments. Han et al. engineered
B. licheniformis to activate dormant pathways for the production of L-alanine at 50 °C, achieving a high yield of optically pure L-alanine (93.7 g/L) [
36]. Ge et al. identified stereospecific dehydrogenases in
B. licheniformis MW3 responsible for the synthesis of 2,3-BDO. They achieved final concentrations of 123.7 g/L for (2R,3R)-2,3-BDO with a productivity of 2.95 g/L/h and 90.1 g/L for meso-2,3-BDO with a productivity of 2.82 g/L/h, both at 50 °C [
37]. By further deleting associated genes, the strain was also able to produce 64.2 g/L of acetoin at 50 °C [
38].
B. licheniformis MW3 can also convert low-cost biomass such as Jerusalem artichoke into meso-BDO, improving efficiency and reducing costs [
29]. Additionally, it can ferment diverse feedstocks, including corn stover and food waste, supporting scalable, eco-friendly biomanufacturing [
39,
40].
Geobacillus thermoglucosidasius (formerly known as
Parageobacillus thermoglucosidus), with optimal growth at 60 °C, is ideal for high-temperature bioproduction. Through metabolic engineering, modified strains produced high levels of L-lactic acid (151.1 g/L) and D-lactic acid (153.1 g/L) at 60 °C with conversion rates of 98.7% and 93.0% [
41]. Additionally, by deleting
ccpN, a transcriptional regulator that redirects glucose metabolism from glycolysis to the pentose phosphate pathway, along with the lactate dehydrogenase genes, the strain produced riboflavin at 1034.5 mg/L within 12 h, underscoring its versatility for high-temperature bio-based chemical production [
42].
A biphasic one-pot biosynthesis system using
Bacillus coagulans and a water/dioctyl phthalate biphasic setup reduced the toxicity of furfural and furfuryl alcohol, achieving a 98% furfural conversion rate and 88% selectivity for furfuryl alcohol within 6 h. This process simultaneously converted glucose to L-lactic acid, producing 264 mM furfuryl alcohol and 64.2 g/L L-lactic acid at 50 °C [
43]. By establishing a reductive TCA pathway and optimizing
pyc expression through promoter screening, biomass was increased, and malic acid production was enhanced to 25.5 g/L, with a glucose conversion rate of 0.3 g/g in biphasic fed-batch fermentation [
44]. Using CRISPR-Cas9 gene editing and adaptive evolution, a high-performance
Lactobacillus paracasei strain was developed, capable of efficiently producing L-lactic acid at 45 °C. In high-glucose open fermentation, it achieved a titer of 221.0 g/L and a productivity of 7.5 g/L/h [
45].
Thermophilic Anaerobic bacteria Thermoanaerobacterium aotearoense SCUT27 has emerged as an efficient host for ethanol production, optimized through genetic modifications that enhance its lignocellulose conversion capabilities [
46,
47]. Knocking out specific redox-related genes to increase intracellular NAD(P)H concentrations and overexpressing the BCD complex to restore growth balance facilitated efficient ethanol production. Fermentation experiments revealed an ethanol production rate of 0.45–0.51 g/L/h and yields ranging from 0.46 to 0.53 g/g sugars at 55 °C [
48]. By introducing sucrose utilization genes and additional genetic modifications,
T. aotearoense SCUT27 demonstrated enhanced growth and metabolic rates using sucrose as a carbon source, improved stress tolerance. It achieved approximately 34 g/L of ethanol in sugarcane molasses. The strain also produced 3.22 g/L of butanol at 55 °C by incorporating genes related to butanol metabolism [
49]. Genetic modifications further enabled SCUT27 to simultaneously metabolize glucose and cellobiose, optimizing its versatility for bioconversion processes [
50,
51].
Clostridium thermocellum is a strictly anaerobic thermophilic bacterium that thrives at temperatures between 45 °C and 65 °C, renowned for its efficient cellulose degradation capabilities and considered a promising host for consolidated bioprocessing (CBP) [52]. Optimization of the isobutanol pathway in this strain yielded 5.4 g/L from 80 g/L cellulose at 50 °C [53]. Through a combination of enzyme and protein engineering, C. thermocellum efficiently produced n-butanol from cellulose, with a yield of 357 mg/L at 55 °C [54]. Expression of chloramphenicol acetyltransferase allowed the biosynthesis of medium-chain fatty acid esters, with total yields of 200 mg/L for various short-chain esters [55]. However, it cannot metabolize xylose naturally, limiting sugar utilization. Introducing xylose metabolism genes from Thermoanaerobacter ethanolicus into C. thermocellum DSM1313 doubled ethanol production efficiency [56].
Thermophilic fungi Myceliophthora thermophila is a thermophilic filamentous fungus known for its rapid cellulose degradation and significant cellulase production, allowing it to effectively break down polysaccharides in plant biomass [
57].
M. thermophila does not exhibit carbon catabolite repression when utilizing cellulose, enabling it to directly transport cellooligosaccharides and xylobiose into the cell. This avoids glucose accumulation, thereby reducing interference with pentose utilization [
58,
59]. To reduce production costs and improve cellulose utilization, researchers overexpressed pyruvate decarboxylase and alcohol dehydrogenase. They modified the bifunctional enzyme PFK-2/FBPase-2 to eliminate FBPase-6 activity, enhancing fructose-2,6-bisphosphate supply and glycolytic rates, which significantly boosted ethanol yield. The optimized strain could directly utilize microcrystalline cellulose and corn husks at 45 °C, producing 52.8 g/L and 39.8 g/L of ethanol, respectively [
60]. Beyond ethanol,
M. thermophila has shown potential in the production of other high-value biochemicals. Gu et al. introduced the fumarase gene from
Candida krusei into
M. thermophila, deleted fumaric acid degradation pathways, and increased the supply of the precursor malic acid. These modifications enabled the engineered strain to produce 17 g/L of fumaric acid from glucose [
61]. The Tianjin Institute of Industrial Biotechnology, in collaboration with Anhui BBCA Group, successfully developed a technology for malic acid production from starch-based sugars. This industrial-scale production line, with a capacity of tens of thousands of tons annually, is now operational. The technology builds on research where
M. thermophila produced malic acid from crystalline cellulose and corn stover at 45 °C, achieving titers of 181 g/L and 105 g/L, respectively [
62].
Thermophilic methylotrophic bacteria One-carbon compounds, including CO
2, methanol, methane, and formic acid, are abundant and cost-effective carbon sources with significant biological utility. They can be effectively converted by methylotrophic microorganisms, thereby reducing greenhouse gas emissions and achieving efficient resource utilization and environmental sustainability [
63].
Bacillus methanolicus is a Gram-positive facultative methylotrophic bacterium capable of growing within a temperature range of 35 °C to 60 °C, with an optimal growth temperature of 50 °C to 55 °C [
64]. By optimizing the expression of pyruvate carboxylase and homoserine dehydrogenase,
B. methanolicus MGA3 can synthesize 69 g/L of L-glutamate and 11 g/L of L-lysine during fed-batch fermentation at 50 °C using methanol as the substrate. Through classical mutagenesis and screening of L-lysine analogs, the mutant strain achieved an L-lysine yield of 65 g/L [
65]. Additionally,
B. methanolicus naturally produces small amounts of riboflavin. Researchers enhanced riboflavin production by overexpressing the riboflavin biosynthesis pathway, achieving a riboflavin titer of approximately 523 mg/L during batch fermentation at 50 °C [
66].
.
Progress in the high temperature metabolic engineering of moderate thermophiles.
| Host |
Temperature |
Carbon Source |
Product |
Titer/Yield |
Reference |
| Bacillus licheniformis |
50 °C |
Glucose |
L-alanine |
93.7 g/L |
[36] |
|
50 °C |
Glucose |
(2R,3R)-2,3-BDO
meso-2,3-BDO
|
123.7 g/L
90.1 g/L
|
[37] |
|
50 °C |
Glucose |
Acetoin |
64.2 g/L |
[38] |
|
50 °C |
Artichoke tubers |
meso-2,3-BDO |
82.5 g/L |
[29] |
|
50 °C |
Corn stover hydrolysate |
2,3-BDO |
74.0 g/L |
[39] |
|
50 °C |
Food waste |
2,3-BDO |
5.9 g/L |
[40] |
| Geobacillus thermoglucosidasius |
60 °C |
Glucose |
L-lactic acid |
151.1 g/L |
[41] |
|
|
|
D-lactic acid |
153.1 g/L |
|
|
60 °C |
Glucose |
Riboflavin |
1034.5 mg/L |
[42] |
| Bacillus coagulans |
50 °C |
Glucose |
L-lactic acid |
64.2 g/L |
[43] |
|
45 °C |
Glucose |
Malic acid |
25.5 g/L |
[44] |
| Thermoanaerobacterium aotearoense SCUT27 |
55 °C |
Lignocellulosic hydrolysates |
Ethanol |
0.46–0.53 g/g |
[48] |
|
50 °C |
Cane molasses |
Ethanol |
34 g/L |
[49] |
|
50 °C |
Cane molasse |
Butanol |
3.22 g/L |
[49] |
| Clostridium thermocellum |
55 °C |
Xylose and cellulose |
Ethanol |
12 mM |
[56] |
|
50 °C |
Cellulose |
Isobutanol |
5.4 g/L |
[53] |
|
55 °C |
Cellulose |
Isobutanol |
5.1 g/L |
[67] |
|
55 °C |
Cellulose |
Ethanol |
29.9 g/L |
[54] |
|
55 °C |
Cellulose |
N-butanol |
357 mg/L |
[54] |
|
55 °C |
Lignocellulosic biomass |
Short-chain sters |
200 mg/L |
[55] |
|
55 °C |
Cellulose |
L-lactic acid |
9.9 g/L |
[68] |
| Myceliophthora thermophila |
48 °C |
Cellobiose |
Ethanol |
11.3 g/L |
[59] |
|
45 °C |
Cellulose |
Ethanol |
52.8 g/L |
[59] |
|
45 °C |
Corncob |
Ethanol |
39.8 g/L |
[60] |
|
45 °C |
Glucose |
Fumaric acid |
17 g/L |
[61] |
| Bacillus methanolicus MGA3 |
50 °C |
Methanol |
L-glutamate |
69 g/L |
[65] |
|
50 °C |
Methanol |
L-lysine |
11 g/L |
[65] |
|
50 °C |
Methanol |
Riboflavin |
523 mg/L |
[66] |
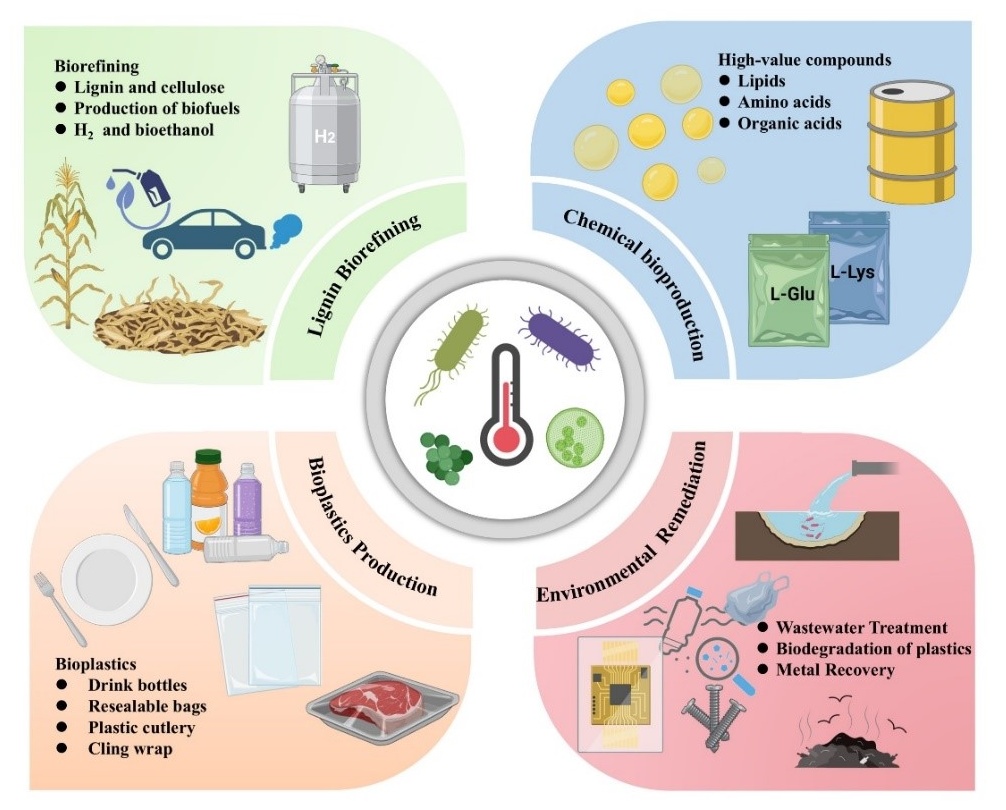
2.3. Sustainable Biorefining and Environmental Applications
2.3.1. Lignin Biorefining
Lignocellulose is the primary component of agricultural and forestry waste, characterized by its dense and complex structure, which hinders its biological degradation and conversion. The development of efficient biocatalysts is crucial for the bioconversion of lignocellulose [
69]. Second-generation biofuel ethanol derived from lignocellulose is an environmentally friendly gasoline substitute, achieving a greenhouse gas emission reduction of up to 115%, significantly surpassing the 19% to 48% reduction associated with food-based ethanol. Therefore, the development of cellulose-derived ethanol is of great importance for addressing food security and energy security issues [
70].
C. thermocellum is an efficient cellulose-degrading bacterium that degrades the polysaccharide components of lignocellulose through the secretion of cellulosomes, a multienzyme complex, demonstrating superior degradation efficiency compared to the free cellulase systems secreted by fungi [
28,
71]. Despite
C. thermocellum’s ability to directly utilize cellulose, its ethanol production has been low. To enhance yields, researchers have employed a functionally complementary microbial co-cultivation strategy. By co-cultivating
C. thermocellum with
Thermoanaerobacter saccharolyticum and blocking the pathways for lactic acid and acetic acid synthesis, the adapted mixed culture achieved an ethanol yield of 38 g/L from Avicel at 55 °C, approaching 80% of the theoretical maximum yield [
72]. This microbial co-cultivation system not only eliminates the need for complex metabolic engineering but also broadens the biorefining platform for lignocellulose, enhancing refining efficiency [
73]. Furthermore, this system has proven effective for the production of succinate and butanol from lignin [
74,
75].
Hydrogen, as a clean energy source, is increasingly recognized for its environmental sustainability throughout production. Research has reported that certain thermophilic anaerobic consortia can utilize untreated cassava mash as a substrate to produce hydrogen gas at 60 °C, achieving H
2 yields of up to 760 mL/L [
76]. Thermophilic anaerobes such as
C. thermocellum and
Thermotoga neapolitana have been employed in the anaerobic fermentation of lignocellulosic biomass for H
2 production [
77].
Thermoanaerobacterium sp. strain F6 is capable of directly utilizing hemicellulosic and cellulolytic materials to generate hydrogen at temperatures as high as 1822.6 mL/L [
78].
2.3.2. Biodegradable Plastics Production
The application of thermophilic microorganisms in the production of biodegradable bioplastics represents a significant breakthrough, as they efficiently convert renewable substrates into valuable bioplastic precursors [
79,
80]. For instance,
Cupriavidus caudatus PHS1 is a strain capable of synthesizing polyhydroxybutyrate (PHA) from various substrates. When cultured with gluconate at 45 °C, this strain accumulated a PHB content of 47% within a dry cell weight (DCW) of 3.14 g/L, approximately 13 times higher than the yield observed at 25 °C [
81]. The strain
Aneurinibacillus sp. H1, isolated from compost, produced 2.03 g/L of poly(3-hydroxybutyrate) from glycerol, accounting for 55.31% DCW at its optimal growth temperature of 45 °C [
82]. The thermophilic bacteria
Weizmannia coagulans MA-13 (formerly
Bacillus coagulans) and
Caldicellulosiruptor sp.DIB 104C has been shown to effectively convert sugars derived from lignocellulose into lactate, a precursor of polylactic acid (PLA), under fermentation conditions at 55 °C [
83,
84,
85].
2.3.3. Lipids Production
Thermophilic microorganisms possess significant potential in lipid production, which are key raw materials for biodiesel production [
86]. Particularly, microalgae are considered ideal hosts for biodiesel production due to their high lipid content and rapid growth rates [
30]. Typically, the lipid content of microalgae comprises 20% to 50% of their dry weight, with some species exceeding 80% lipid content [
87,
88]. Thermophilic microalgae have an optimal growth temperature that exceeds the typical range of 16–27 °C for regular microalgae, often around 50 °C. Furthermore, thermophilic algae have a significant advantage in biodiesel production due to their heat-stable enzymes, which maintain activity at high temperatures, thereby enhancing their potential in biofuel production [
89]. A study comparing the lipid content of
Gloeocapsa gelatinosa, isolated from the hot spring “Aïn Echfa” at 60 °C, revealed that high light intensity combined with high temperature (60 °C) and the lowest NaNO
3 concentration increased the proportion of saturated fatty acids 16:0 and 18:0 [
90].
Chlorella sorokiniana can tolerate high temperatures of up to 42 °C, with an optimal growth temperature of 37 °C. Under these conditions, its maximum lipid content reached 31.5%, and the lipid productivity reached 2.9 g/L/d [
91]. These findings make thermophilic microalgae promising candidates for sustainable biofuel production.
2.3.4. Environmental Remediation
Biodegradation of plastics The use of thermophilic microorganisms for plastic degradation in contaminated thermal habitats offers significant advantages, as high-temperature environments alter the physical and optical properties of plastics, enhancing their bioavailability and solubility [
92]. Through metagenomic analysis, researchers identified
Brevibacillus thermoruber in plastic-polluted geothermal areas, which exhibited high degradation activity against poly-ε-caprolactone (PCL) at 55 °C, accounting for 63.6% of the total microbial community’s degradation capacity [
93].
Streptomyces thermoviolaceus can secrete extracellular PCL depolymerases, achieving complete degradation of 0.1% of the substrate after 6 h at 45 °C [
94]. A thermophilic whole-cell catalyst capable of degrading polyethylene terephthalate (PET) was constructed by expressing heterologous thermophilic cutinase in
C. thermocellum. Significant degradation of commercial PET films was observed at 60 °C, with more than 60% of the initial mass (approximately 50 mg) being degraded. [
95].
Bacillus pallidus, a thermophilic bacterium, was able to degrade nylon 12 and nylon 6 at 60 °C [
96].
Wastewater Treatment Thermophilic microalgae exhibit significant potential for applications in wastewater treatment. They effectively remove nutrients such as nitrogen, phosphorus, heavy metals, and organic compounds while converting biomass into energy and materials [
97]. They can directly grow in the hot wastewater upon discharge, eliminating the need for a cooling step and simplifying subsequent wastewater treatment. Additionally, the elevated temperature accelerates chemical reaction rates, significantly enhancing pollutant degradation efficiency and making the treatment process more efficient compared to traditional methods [
98]. For example,
G. sulphuraria can grow at 56 °C, showing promise in treating wastewater derived from food waste hydrolysates [
99,
100]. In primary settling wastewater,
G. sulphuraria significantly reduced the concentrations of organic carbon, ammonium, and phosphates, achieving removal rates of 46–72%, 63–89%, and 71–95%, respectively [
101]. Furthermore, the bacterial count in the treated wastewater is markedly decreased, with the complete elimination of
Enterococcus faecalis and
E. coli [
102].
Chlorella thermophila, a thermophilic microalga, has been cultivated in nutrient-rich dairy wastewater, effectively reducing organic carbon and nutrient loads, while the extracted biomass demonstrated the ability to inhibit the growth of plant pathogens [
103]. Co-culture systems, such as
Streptomyces thermocarboxydus in conjunction with
Chlorella sorokiniana, facilitated biodiesel production while treating cassava biogas wastewater, with biomass and lipid yields increasing by 21% and 25%, respectively [
104].
Metal Recovery In metal bioremediation, microorganisms stabilize or transform metal ions through enzymatic or non-enzymatic oxidation/reduction reactions, thereby reducing their ecological risks [
105]. Additionally, microbial surface adsorption and the secretion of metabolites such as amino acids, proteins, and peptides can precipitate with metal ions, further stabilizing them [
106]. Thermophilic microorganisms exhibit unique advantages in exothermic bioleaching processes, as they not only adapt to high-temperature environments but also mitigate mineral surface passivation, thereby enhancing metal recovery efficiency [
107]. Through mixed cultures of thermophilic bacteria, effective biolixiviation of pretreated waste printed circuit boards (PCBs) has been achieved, with recovery rates for Zn, Cu, and Al reaching 85.23%, 76.59%, and 70.16%, respectively, within seven days. Within this community, two thermophilic bacterial species,
Leptospirillum ferriphilum and
Acidithiobacillus caldus, were identified and both exhibited robust performance in the bioleaching process [
108]. In another study, thermophilic microbial communities were employed for the biolixiviation of various metals from contaminated sediments. The co-culture system of
Sulfobacillus thermotolerans and
Acidithiobacillus caldus demonstrated superior acidification efficiency and redox potential, significantly enhancing the bioleaching of sulfur and pyrite.
S. thermotolerans further reduced toxicity by oxidizing leachate byproducts, facilitating the bioleaching process. Compared to monocultures, the co-culture exhibited higher proliferation rates and cell densities, further validating the synergistic effect and contributing to improved bioleaching efficiency [
109]. These indicated that thermophilic microorganisms not only enhance metal recovery efficiency in biolixiviation but also play a crucial role in environmental remediation, particularly in the treatment of industrial wastewater and sludge contaminated with heavy metals.
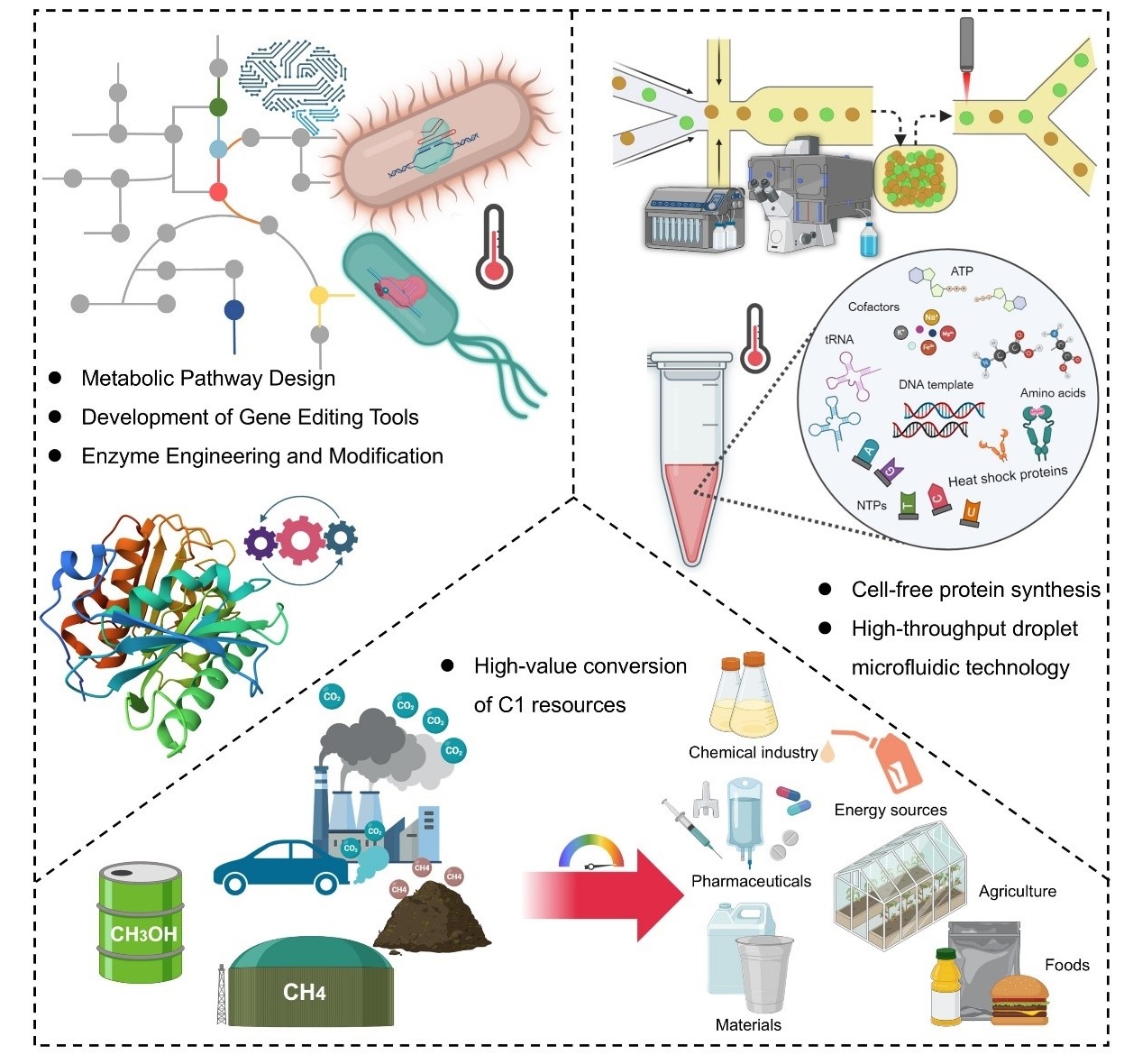
2.4. Acting as the Evolving Platform of Thermophilic Enzymes
2.4.1. Co-Evolution of Thermophilic Enzymes and Thermophiles
Thermophiles employ a range of adaptive mechanisms to survive and reproduce in high-temperature environments. These adaptations are closely linked to the co-evolution of thermozymes, ensuring cellular functions such as DNA synthesis, protein expression and cell metabolism remain stable and efficient under extreme conditions [
110].
Thermophilic bacteria enhance the stability of their cell membranes by increasing the proportion of saturated fatty acids, extending phospholipid alkyl chain lengths, and incorporating more isomerized branched chains. These optimizations not only improve the rigidity and stability of the membrane but also provide a more stable cellular environment for enzymes, ensuring their structures and functions [
111,
112]. High temperatures pose significant threats to DNA integrity, driving thermophilic microorganisms to evolve stable nucleic acid structures and efficient DNA repair systems, where thermally stable repair enzymes have co-evolved to indirectly ensure the accuracy of protein synthesis [
113]. For instance, the DNA-guided Argonaute protein TtAgo in
Thermus thermophilus aided in the replication of the circular genome and conferred a competitive advantage to the organism [
114]. Additionally, the genomes of thermophiles contain specific enzymes that induce negative supercoiling to maintain DNA stability [
115,
116]. Heat shock proteins (HSPs) protect intracellular proteins from misfolding or aggregation, inhibit cascades of cell death signaling, and preserve intracellular signaling pathways essential for the cell viability of thermophiles [
117,
118].
Methanosarcina thermophila TM-1 possesses an active Hsp70 chaperone machine that induces a heat shock response, along with the stress gene trkA, which exhibits induction activity similar to classic heat shock genes [
119]. The expression of heat shock protein-related genes has been enhanced at high temperatures to facilitate proper protein folding and maintain functionality [
120,
121].
Enzymes have gradually evolved to maintain activity and stability at elevated temperatures through mutation accumulation and natural selection, thereby supporting cellular survival in extreme environments [
122,
123]. Firstly, thermozymes have evolved their amino acid composition and structural characteristics to maintain stability at high temperatures. Compared to mesophilic proteins, thermophilic enzymes contain a higher number of charged amino acids, which stabilize the protein structure through electrostatic interactions [
124,
125]. Aromatic amino acids further enhance protein stability through cation-π interactions [
126,
127,
128]. Secondly, the intramolecular interactions of thermozymes have also evolved, including both covalent bonds and non-covalent forces [
129,
130,
131]. For example, strong electrostatic and hydrophobic interactions prevent aggregation and enhance the stability of thermophilic proteins [
37,
132]. Though high temperatures destabilize proteins by increasing entropy, disulfide bonds between cysteines stabilize protein structure [
133,
134,
135]. Moreover, thermophilic proteins are more rigid than mesophilic proteins, providing stronger structural support [
136,
137]. Elevated proline content also enhances protein rigidity and thermal stability, while glycine reduces the stability due to high entropy [
138,
139,
140,
141,
142]. Extreme thermophilic proteins minimize destabilization from solvation by burying hydrophobic surface area [
143,
144].
2.4.2. Serving as the High-Value Reservoir of Thermozymes
Thermophilic bacteria are considered a valuable reservoir of thermozymes. To date, various thermozymes have been characterized, such as xylanases, proteases, cellulases, amylases, and lipases [
11]. These enzymes exhibit high catalytic activity at elevated temperatures, offering advantages such as ease of expression in mesophilic hosts, enhanced substrate solubility and reduced viscosity for improved reaction efficiency, minimized microbial contamination, and increased resistance to chemical denaturants, making them ideal for complex industrial applications [
12]. Thermozymes have achieved numerous successful applications in commercial industries. For example, Lipozyme
® TL IM, a thermophilic lipase derived from
Thermomyces lanuginosus and developed by Novozymes, is widely used in the stereoselective hydrolysis and transesterification of esters, while the company’s thermostable laccase product, Denilite
® IIS, is commonly employed in textile industries for processes such as fabric bleaching [
145,
146]. ASA Spezialenzyme GmbH has developed Xylanase 2X
®, sourced from
Trichoderma sp., primarily for the degradation of cellulose [
147]. These applications highlight the tremendous potential and broad utilization of thermophilic enzymes across various industrial sectors. Representative thermozymes with potential application values are listed in .
Thermophilic lipases, characterized by their exceptional lipid solubilization capacity, have extensive applications in detergents, the paper industry, and biodiesel production. These enzymes have been identified from a wide range of bacterial sources, including
Pseudomonas,
Bacillus, and
Acinetobacter, highlighting their versatility and industrial significance [
148]. For example, a highly thermostable lipase has been isolated from
Bacillus stearothermophilus, with a remarkable half-life of 15–25 min at 100 °C and an optimal catalytic temperature of 78 °C [
149]. Similarly, lipases derived from
Pseudomonas moraviensis and
Thermosyntropha lipolytica exhibit optimal enzymatic activity at 65 °C and 96 °C, respectively, underscoring their potential for high-temperature industrial applications [
150,
151].
Chitinase is capable of converting insoluble chitin into bioactive chito-oligosaccharides and monomers, and degrading the chitin exoskeleton or cell walls of pathogens to inhibit their growth, thereby playing an important role in the skincare industry and biological control [
152]. The chitinase from
B. licheniformis MB-2 performs optimally at 70 °C [
153]. Cellulase and amylase have been applied in the food industry and the paper industry, where they enhance fiber softness and uniformity, thereby improving paper quality [
154]. A thermophilic amylase (PaAFG) screened out from the hyperthermophilic archaeon
Pyrococcus abyssi showed maximum activity at 90 °C and exceptional thermal stability between 60–100 °C [
155].
In the field of molecular biology, thermozymes are highly valued for their ability to maintain activity at elevated temperatures. A notable example was the DNA polymerase, which has been isolated from the thermophilic bacteria such as T
hermus aquaticus and
Thermococcus barophilus [
156,
157,
158]. Additionally, DNA ligases have also been isolated and characterized from
Thermococcus sp. 1519,
Pyrococcus furiosus, and
Staphylothermus marinus [
159,
160,
161]. The DNA ligase derived from
Thermococcus sp. 1519 exhibited optimal activity at 60–70 °C and was capable of ligating DNA fragments with long overhangs, making it a promising tool for Gibson assembly application [
159].
.
Representative thermozymes with potential applications in industry and scientific research.
| Scenario |
Thermozymes |
Optimal Temperature |
Application |
Organism |
Reference |
| Foods |
α-amylase |
90 °C |
Enhancing dough fermentation in baking; Advancing product quality, fermentation efficiency, and clarity in the brewing industry. |
Pyrococcus abyssi |
[155] |
| Xylanase |
70–75 °C |
Myceliophthora thermophila |
[162] |
| Cellulase |
102–105 °C |
Pyrococcus furiosus |
[163] |
| Pectinases |
90–100 °C |
Thermotoga maritima |
[164] |
| Mannanase |
60 °C |
Reducing viscosity, enhancing texture, and improving beverage clarity. |
Aspergillus fumigatus HBFH5 |
[165] |
| Personal and household care products |
α-amylase |
90 °C |
Improving paper quality and enhancing bleaching processes in the paper industry. |
Pyrococcus abyssi |
[155] |
| Cellulase |
75 °C |
|
Caldicellulosiruptor bescii |
[166] |
| Pectinases |
90–100 °C |
|
Thermotoga maritima |
[164] |
| Lipases |
70 °C |
|
Geobacillus stearothermophilus |
[167] |
| Laccase |
90 °C |
|
Chitinophaga sp. CB10 |
[168] |
| Glucoamylases |
75 °C |
Effectively breaking down starch-based stains for enhanced detergent performance. |
Thermoanaerobacter ethanolicus |
[169] |
| Proteinase |
60 °C |
Effectively removing blood, milk, and similar stains to enhance detergent performance. |
Thermus thermophilus HB8 |
[170] |
| Esterase |
60 °C |
Breaking down grease and oil stains to enhance detergent performance. |
Geobacillus thermodenitrificans NG80-2 |
[171] |
| Lipases |
70 °C |
Geobacillus stearothermophilus |
[167] |
|
78 °C |
|
Fervidobacterium changbaicum CBS-1 |
[149] |
| Lipases |
65 °C |
Biodiesel production. |
Burkholderia ubonensis SL-4 |
[172] |
|
Chitinase |
90 °C |
Production of chitosan for applications in moisturizing skincare products. |
Thermotogae bacterium |
[173] |
| Environmental protection |
Esterase |
80 °C |
Decomposing ester compounds in wastewater or pesticides. |
Thermoanaerobacterium thermosaccharolyticum DSM 571 |
[174] |
| Laccase |
60 °C |
Removing phenolic contaminants from industrial wastewater; degrading pesticide residues. |
Geobacillus yumthangensis |
[175] |
| Chitinase |
45 °C |
As a biocontrol agent. |
Streptomyces alfalfae ACCC 40021 |
[176] |
| Molecular biology |
DNA polymerases |
80 °C |
PCR. |
Thermus aquaticus |
[156] |
|
72–74 °C |
Thermus scotoductus K1 |
[177] |
| Ligases |
70 °C |
Ligase chain reaction. |
Thermococcus sp. 1519 |
[160] |
2.5. High-Temperature Whole-Cell Catalysis
Whole-cell biocatalysts (WCBs) utilize the entire cell to carry out catalytic reactions, eliminating the requirement for additional cofactors and the processes of enzyme purification [
178]. These systems are highly stable and reproducible, but low production efficiency remains a challenge. For example, industrial strains like
E. coli grow optimally at 37 °C, yet the complexity of intracellular enzymes and metabolite interactions limit the efficiency [
34]. Elevated temperatures simplify the metabolism by reducing side reactions and byproduct formation, enabling high-selectivity whole-cell biocatalysts with yields comparable to purified enzymes [
179].
Researchers have identified hydrogen-dependent CO
2 reductases (HDCRs) in the thermophilic acetic acid bacteria
Acetobacterium woodii and
Thermoanaerobacter kivui, which efficiently catalyzed the conversion of CO
2 to formic acid. Leveraging this enzyme, they developed a whole-cell biocatalyst to convert H
2 and CO
2 into formic acid, showing potential applications in hydrogen storage and CO
2 capture [
180]. Additionally, a thermally stable lipase from
Geobacillus thermocatenulatus (BTL2) was introduced into
Aspergillus oryzae to generate an immobilized whole-cell biocatalyst, which retained high activity at 60 °C. This catalyst effectively converted palm oil into biodiesel, achieving conversion rates close to 100% at elevated temperatures between 40 °C and 50 °C [
181].
There have been reports of WCBs utilizing thermozymes expressed in non-thermophilic bacteria. For instance, by expressing the thermophilic lipase gene from
Rhizobium etli in
E. coli, researchers catalyzed the ring-opening polymerization of ε-caprolactone, achieving nearly 100% monomer conversion at temperatures between 70 °C and 90 °C. Even after 10 reuse cycles, the monomer conversion rate remained above 90% [
182]. Additionally, Peng et al. employed bioinformatics techniques to identify thermophilic
N-acetyl-D-glucosamine epimerase (AGE) and
N-acetylneuraminic acid aldolase (NanAs) that remain stable and exhibit high enzymatic activity at 60 °C. Utilizing the rapidly growing
Vibrio natriegens, they established a high-temperature WCB producing
N-acetyl-D-neuraminic acid (Neu5Ac) at an average rate three times higher than that at 37 °C, achieving titers of 126.1 g/L and a productivity of 71.6 g/(L·h), which is 7.2 times higher than that of
E. coli [
34].
The integrity of the cell membrane in non-thermophilic bacteria like
E. coli is compromised at elevated temperatures. This limits the continuous reuse of whole-cell biocatalysts. It is crucial to enhance the efficiency of high-temperature WCB by stabilizing either the enzymes or the cell structure [
182]. To address this, glutaraldehyde treatment was applied to
E. coli expressing thermophilic fumarase from
Thermus thermophilus, stabilizing the system without altering cell structure [
179]. Given that circular proteins exhibit greater thermal stability and structural integrity than linear proteins, enzyme cyclization using SpyTag and SpyCatcher has been used to enhance the thermal tolerance and activity of β-glucosidase, improving catalytic efficiency and retaining 50% activity after 10 days of storage at room temperature [
183].
The utilization of thermozymes is highly significant in scientific research, industrial applications, and environmental protection. Studying thermozymes enhances our understanding of the molecular mechanisms behind enzyme evolution and protein thermal stability, laying the groundwork for advancements in protein engineering and molecular biology [
184]. The bioprospecting and engineering of these enzymes further promote the development of innovative biotechnologies that greatly improve the efficiency of biocatalysts, which is invaluable for advancing industrial biotechnology and synthetic biology. However, most enzymes lack heat resistance above 50 °C, making the discovery of thermally stable enzymes and the improvement of protein thermal stability important topics in scientific research and industrial applications [
185]. Here, we discuss cases of thermophilic enzyme exploration and modification guided by computational methods, artificial intelligence (AI), and high-throughput screening techniques. Representative tools for thermozymes discovery and enzyme thermal stability optimization are listed in .
3.1. Bioinformatics-Guided Bioprospecting of Thermozymes
Enzyme thermal stability is crucial for extending reaction duration and facilitating reusability. Traditional methods for isolating thermozymes typically rely on the extraction from bacteria or archaea found in high-temperature environments. However, these wet-lab processes are relatively expensive and time-consuming. Rapid advancements in molecular biology, bioinformatics, and genomics facilitate the use of sophisticated bioinformatics tools and omics data to discovery novel enzymes.
Co-evolution Mining The co-evolution of enzymes and thermophilic microorganisms allows the identification of thermozymes from the thermophilic genomes. To this end, a co-evolutionary platform, Co-Evolution Mining (CEM), has been developed for thermozymes discovery. Using glucosamine-6-phosphate acetyltransferase (GNA1) from
Saccharomyces cerevisiae as a template, 1672 microbial genomes with T
opt over 50 °C and high GC content were analyzed. Four novel GNA1 enzymes with excellent stability have been identified, enabling the production of N-acetylglucosamine (GlcNAc) at 119.3 g/L [
186].
Omics-Based Bioprospecting Protein-coding genes with potential thermophilic properties could be identified utilizing genomic databases by comparing existing thermophilic enzyme sequences. Using reported heat-stable ω-transaminases as templates, a novel heat-stable ω-transaminase was identified from thermophilic microbial genomic databases, retaining nearly unchanged activity below 80 °C and exhibiting typical thermophilic traits [
187]. A GeneBank-mining virtual probe to discover thermophilic glucose isomerases has been designed using energy calculations from FoldX and Rosetta. This probe facilitated the identification of two thermophilic enzymes, which retained 40.6% and 52.6% relative residual activity, respectively, after incubation at 90 °C for 24 h [
188]. Using Tagatose 4-epimerase UxaE from
T. neapolitana as a virtual probe and performing protein clustering, Thar-T4Ease from
Thermoprotei archaeon was identified, exhibiting remarkable thermostability with a half-life of 198 h at 80 °C and an 18.9% conversion rate using 100 g/L D-fructose as the substrate [
189]. By performing length screening and clustering analysis on 358 sequences from the NCBI database, in combination with structural predictions and substrate docking using AlphaFold, the D-Allulose 3-epimerase (DAEase) from
Thermogutta terrifontis was identified, exhibiting a half-life of 32 h at 70 °C [
190].
Metagenomics, also known as environmental genomics or community genomics, effectively overcomes the limitations of traditional microbial cultivation methods and has been widely applied to discover novel genes [
191,
192,
193]. For instance, wastewater metagenomes from tanneries have been analyzed and machine learning-based regression models have been used to predict the optimal pH and temperature of lipases, characterizing a newly thermophilic lipase showing maximum activity at 60 °C [
194]. Plant composting is a process in which thermophilic microorganisms degrade plant-based polymers. Seven enzymes with polyester hydrolytic activity have been identified from the metagenomic samples from a composting facility, and enzyme PHL7 completely hydrolyzed untreated PET plastic waste at a rate of 0.6 mg/g PET within 24 h at 70 °C [
195]. Sun et al. used transcriptomics to analyze protein transcription levels at different composting stages and identified a chitinase, ActChi, which was active at 80 °C. The catalytic activity was further enhanced by 400% by incorporating a hyperthermophilic chitin-binding domain [
196].
.
Tools for thermozymes discovery and thermal stability optimization.
| Tools |
Functions |
Input Type |
Key Methods |
URL |
Reference |
| TOME |
Predicting the Topt |
Sequence |
SVR |
https://github.com/EngqvistLab/Tome (Accessed on 6 January 2025) |
[197] |
| TOMER |
Predicting the Topt |
Sequence |
SVR |
https://github.com/jafetgado/tomer (Accessed on 6 January 2025) |
[198] |
| Preoptem |
Predicting the Topt |
Sequence |
FNN; CNN |
http://www.elabcaas.cn/pird/preoptem.html (Accessed on 6 January 2025) |
[199] |
| Pythia |
Predicting the ΔΔG |
PDB structure |
MPNN |
https://pythia.wulab.xyz/ (Accessed on 6 January 2025) |
[200] |
| FoldX |
Predicting the ΔΔG |
PDB structure |
Energy calculation |
https://foldxsuite.crg.eu/ (Accessed on 6 January 2025) |
[201] |
| ROSETTA |
Predicting the ΔΔG |
PDB structure |
Energy calculation |
https://rosettacommons.org/ (Accessed on 6 January 2025) |
[202] |
| FireProt2.0 |
Predicting the ΔΔG and Tm |
Sequence/PDB structure |
Bron-Kerbosch |
https://loschmidt.chemi.muni.cz/fireprotweb/ (Accessed on 6 January 2025) |
[203] |
| DeepSTABp |
Predicting the Tm |
Sequence |
Transformer |
https://csb-deepstabp.bio.rptu.de/ (Accessed on 6 January 2025) |
[204] |
| BayeStab |
Evaluating the impact of single mutations on protein stability |
PDB structure |
GNN |
http://www.bayestab.com. (Accessed on 6 January 2025) |
[205] |
| GeoStab-suite |
Prediction of protein mutation fitness scores, ΔΔG, and ΔTm |
PDB ID |
GAT; MLP |
https://structpred.life.tsinghua.edu.cn/server_geostab.html (Accessed on 6 January 2025) |
[206] |
| HotSpot Wizard3.0 |
Identifying hotspots for enhancing enzyme thermal stability |
Sequence |
Energy calculation |
http://loschmidt.chemi.muni.cz/hotspotwizard (Accessed on 6 January 2025) |
[207] |
| ProThermDB |
Database of protein thermal stability |
Sequence |
/ |
https://web.iitm.ac.in/bioinfo2/prothermdb/index.html (Accessed on 6 January 2025) |
[208] |
| BRENDA |
Database of enzymes’ optimal temperature |
EC Number/Name of the Gene or Enzyme |
/ |
https://www.brenda-enzymes.org/ (Accessed on 6 January 2025 ) |
[209] |
| ThermoMutDB |
Database of protein thermal stability |
Protein Name/PDB ID |
/ |
https://biosig.lab.uq.edu.au/thermomutdb/ (Accessed on 6 January 2025) |
[210] |
3.2. High-Throughput Screening of Thermostable Enzymes
Traditional protein engineering is primarily achieved through rational or semi-rational design and directed evolution techniques. Directed evolution in combination with high-throughput screening method does not require an understanding of protein structures, allowing for the direct identification of advantageous mutations [
211]. The high-throughput screening method plays an important role in the evaluation of enzyme activity, stability, and other key performance characteristics, as well as addressing the time and cost challenges associated with multiple rounds of iterative screening. Rational design of enzymes relies on a solid foundation of protein structures and amino acid mutations [
212]. Semi-rational design involves the construction of mutant libraries through multiple rounds of mutagenesis, followed by iterative screening of beneficial variants [
213]. AI-driven virtual screening approaches combining computational tools and big data analysis present an efficient pathway for enzyme engineering, although experimental validation remains essential.
3.2.1. High-Throughput Screening
The high-throughput screening method allows for the quantitative analysis of substrates or target products by measuring the changes in absorbance or fluorescence. For instance, Li et al. developed a high-throughput screening system for D-allulose 3-epimerases (DAEases) by measuring the product D-allulose via determimning NAD
+ absorbance at 340 nm, identifying a mutant with 1.2-fold higher activity and 8.7 °C improved thermal stability, compared to the wild-type enzyme’s T
5060 value of approximately 70 °C. [
214]. To withstand the high temperatures encountered in PET processing, Shi et al. developed a high-throughput screening method to monitor PET hydrolase activity using bis (2-hydroxyethyl) 2-hydroxyterephthalate (BHET-OH) as a novel substrate, producing the fluorescent product and yielding an optimal mutant with a T
m increase of 23.3 °C, from the original T
m of 46.13 °C to a final T
m of 69.38 °C [
215]. Agar plate screening technology is useful in assessing enzyme activity through hydrolysis halos or colorimetric reactions. Zhou et al. optimized the agar plate material with improved heat resistance, identifying a hyperthermostable polyphosphate glucokinase mutant with a 7200-fold increase in half-life and a T
m increase of 19.8 °C, from the original T
m of 55.9 °C to a final T
m of 75.7 °C [
216]. This method has also been extended to screening thermophiles. Xiong et al. used Congo red agar and colorimetric assays to isolate
Streptococcus thermophilus mutants, leading to a 25.42% increase in EPS production [
217].
Biosensors are essential in synthetic biology for monitoring microbial behavior, metabolism, and product accumulation. Commonly based on transcription factors or riboswitches, they respond by changing protein or nucleic acid conformation, triggering gene expression, and producing detectable signals like fluorescence, growth rates, or pathway expression levels [
218,
219]. Biosensors enable rapid evaluation of enzyme activity, stability, and substrate affinity, crucial for directed evolution and high-throughput screening [
220]. Using a biosensor specifically responsive to D-allulose, researchers identified DAEase variants with enhanced catalytic efficiency and thermal stability, achieving a half-life 17.23 times longer than the wild type at 65 °C [
221]. Similarly, Li et al. utilized a D-allulose-responsive biosensor to design an in vivo growth-coupled screening platform, significantly improving the thermal stability of ketose 3-epimerase (KEase) through directed evolution [
222]. Biosensors that reassemble split GFP fragments to produce fluorescence upon ligand binding have been used for assessing enzyme stability and solubility [
223]. Leveraging this type of biosensor, researchers have identified the mutant of thermostable esterase Aaeo1, I51V-E170D, that possesses 4.5-fold higher activity than the wild type [
224].
3.2.2. AI-Driven Prediction and Iterative Virtual Screening
AlphaFold is a revolutionary tool utilizing deep neural networks, integrating sequence data from known structures, co-evolutionary information, and distance and angle data between amino acids, to predict the three-dimensional (3D) structure of target proteins through iterative training and testing [
225,
226]. The combination of AlphaFold2 and Rosetta methods significantly enhances the accuracy of enzyme thermal stability predictions. For example, a computational protocol using AlphaFold and Rosetta integrated global conformational sampling with energy predictions, improving the prediction of enzyme thermal stability substantially [
227]. A Co-Evolution Mining (CEM) tool has been developed to search for thermophilic NanA, and 92 proteins with structures predicted by AlphaFold2 were subjected to molecular docking with GlcNAc and pyruvate [
34]. Further assessments of protein stability and aggregation propensity using FoldX and Aggrescan3D identified the N-acetylneuraminic acid aldolase (AGE) from
Thermophagus xiamenensis as the best candidate, which exhibited an optimal temperature of 65 °C and retaining over 50% of its initial activity after 60 h of treatment. Wang et al. designed an Ancestral Sequence-Structure-Molecular Dynamics (ASSMD) strategy to explore ancestral enzyme sequences [
228]. They established a comprehensive structure library for ancestral nitrilase enzymes by predicting their structures by AlphaFold 2.0. Additionally, they generated a dynamic parameter library that revealed the biochemical properties of these enzymes via molecular dynamics simulations and provided insights into the structural rigidity of the protein dynamics by analyzing the Root Mean Square Deviation (RMSD). Notably, they identified an ancestral enzyme ASR135 remaining with the residual enzymatic activity at 11.69% after incubation at 90 °C for 10 min. The laboratory evolution further enhanced the thermal stability of ASR135, yielding the mutant ASR135-M4 retaining hydrolytic activity even at 100 °C.
The implementation of AI-driven virtual screening method leverages cloud platforms to perform massive computations and cluster architecture calculations, enabling iterative virtual screening of thermostable enzymes [
229]. For example, sequence similarity-based annotations and supervised learning algorithms have been employed to characterize cellulose-degrading enzymes from high-throughput metagenomic data. Through iterative cross-validation testing, a heat-stable cellulose-degrading enzyme was successfully identified [
230]. Zhang et al. developed a deep learning tool, Preoptem, to screen thermophilic proteins from a marine metagenome and predicted 23.82% of 3199 proteins were thermophilic, leading to the discovery of a thermophilic chitinase, Chi304, with a Tm of 89.65 ± 0.22 °C [
199]. AI-driven iterative virtual screening is also applicable to the screening of enzyme mutants. Xi et al. performed virtual saturation mutagenesis on the dimeric interface residues of L-lysine decarboxylase (CadA), in combination with computationally guided virtual screening, obtaining an optimized mutant with enhanced stability under high-temperature and high-pH conditions, and increasing cadaverine production by 37% at 50 °C and pH 8.0 [
231]. Similarly, Li et al. applied a dual-focusing strategy combining hierarchical clustering and iterative screening to develop thermostable methylphosphonic acid hydrolase (MPH), obtaining a mutant with a 13.3 °C T
m increase and 4.2-fold higher catalytic activity [
232]. Qi et al. employed structure-guided virtual screening and a combinatorial active site saturation testing/iterative saturation mutagenesis custom strategy to engineer D-allulose 3-epimerase (DAEase) from
T. terrifontis, obtaining a mutant (P105A/L14C/T63G/I65A) displaying a 5.12-fold increase in catalytic activity [
233].
3.3. AI-Guided Design and Engineering of Thermostable Enzymes
AI is crucial in identifying, optimizing, and designing thermally stable enzymes [
234]. By analyzing structural, mutation, and experimental data, AI predicts stability changes and guides protein engineering, reducing experimental reliance [
235,
236]. Integrating AI with bioinformatics enhances our understanding of enzyme behavior at high temperatures, advancing enzyme development for industrial and biotechnological applications [
237]. depicts the general workflow of using machine learning to predict and enhance enzyme thermal stability. First, data processing involves collecting and extracting key features such as T
m and T
opt to prepare for subsequent analysis. Next, the modeling phase uses neural networks to build a predictive model for enzyme thermostability. Finally, the prediction and design step applies the model to evaluate enzyme performance and optimize critical sites through hotspot design, thereby enhancing enzyme stability under high temperatures.
Machine learning techniques have been utilized to enhance the thermal stability of pectate lyase PMGL-Ba from
Bacillus stearothermophilus by training and optimizing various amino acid features of 152 mutants. Among the six regression algorithms, the Support Vector Regression (SVR) model performed the best in predicting thermal stability. Using this model, the mutant P36 exhibited a significantly improved half-life at 75 °C and 80 °C and an increased activity by 2.1 times was characterized [
238]. Yoshida et al. developed a loop-walking method to improve protein thermal stability by introducing random mutations into three consecutive amino acid residues within each loop. The critical loop region L7 was characterized using a discriminative model with the physicochemical parameters of each amino acid position as input and the thermal stability of 214 mutants as output. This approach identified the mutant P233D/L234P/V235S, which retained 66% activity after heat treatment at 60 °C, significantly higher than the wild-type enzyme (5%) [
239].
Deep learning models use multi-layer neural networks to handle and analyze vast amounts of protein structural and sequence information [
240]. For example, MutCompute24 is an algorithm developed using a three-dimensional (3D) self-supervised convolutional neural network (CNN). It learns variations in the local microenvironment of amino acids from extensive structural information in the PDB database to determine whether structural changes are beneficial for protein stability [
241]. Lu et al. employed this model to predict mutations that enhance the thermal stability of the plastic-degrading enzyme PETase, finding that 85% of 27 potential mutation combinations exhibit greater thermal stability compared to the starting enzyme [
242]. A CNN algorithm, developed using a dataset of single amino acid mutation data (S3661) selected from the ProTherm protein thermodynamic database, has been demonstrated to be more accurate than traditional methods in predicting the thermal stability of mutants of
Rhizomucor miehei lipase [
243]. Liao et al. introduced the concept of the protein bottleneck effect, highlighting that the thermal stability of a protein is constrained by its least stable region. Using α-Amylase as a model, they identified its B domain as the critical region. They then developed the Zero-Shot Hamiltonian (ZSH) model, based on the MSA Transformer protein language model, to predict thermal stability changes from double mutations, successfully identifying a mutant F237R/S240G with an 8.5 °C increase in T
m [
244].
. General workflow of machine learning-assisted protein thermostability design.
Thermophilic microorganisms, due to their robust growth and contamination resistance at high temperatures, are prime candidates for NGIB chassis. These microbes boost biomanufacturing efficiency and sustainability, offering novel strategies in biofuels, biocatalysis, and environmental cleanup. Their enzymes have wide applications, and researchers have harnessed thermozyme sequences and structures to improve mesophilic enzyme stability. This review examines the advances of thermophiles and thermozymes, highlighting their significant roles as high-temperature catalysts in NGIB. It also highlights advanced technologies like AI and high-throughput screening technologies in thermozyme discovery and development, along with the economic and environmental benefits of thermophilic operations. The review demonstrates that advances in these fields could significantly enhance industrial biocatalysis, supporting a more efficient and eco-friendly bioeconomy aligned with global carbon neutrality goals.
Our experiments related to this review were carried out in Biological & Medical Engineering Core Facilities of Beijing Institute of Technology.
J.L.: Investigation, Writing—original draft, Visualization, Writing—review & editing. L.S.: Conceptualization, Writing—original draft, Writing—Review & Editing, Project administration. Y.-X.H.: Conceptualization, Writing—Review & Editing, Project administration.
Not applicable.
Not applicable.
Not applicable.
This work was supported by Hebei Natural Science Foundation (C2023105022), Science and Technology Program of Tangshan (23130228E), the Beijing Institute of Technology Research Fund Program for Young Scholars, and the BIT Research and Innovation Promoting Project (Grant No. 2024YCXY048).
The authors declare no competing financial interest.