1. Introduction
Plastic is one of the most commonly produced synthetic materials globally, playing a significant role in industries like packaging, construction, automotive, electronics, and consumer goods. In 2019, nearly 460 million tons of plastic products were produced worldwide, bringing the cumulative total to over 8 billion tons [
1,
2]. Plastics have largely substituted traditional materials like metal and wood, yet the disposal of plastic waste continues to be a major challenge. In 2022, around 380 million tons of plastic waste were produced, primarily from common plastics like polyethylene terephthalate (PET), polyvinyl chloride (PVC), and polyethylene (PE), due to inadequate recycling and waste management systems [
3]. However, the chemical stability of these plastics makes them highly durable and resistant to natural breakdown [
4]. Furthermore, about 8 million tons of plastic enter the ocean annually, leading to marine animal deaths from consuming large quantities of plastic particles. These plastic particles can accumulate in humans through seafood, sea salt, and other pathways, ultimately threatening human [
5,
6]. Developing recycling technologies for waste plastic resources is a crucial approach to addressing the “white pollution” issue ().
. Overview of the Biodegradation and upgrading of Waste Plastic.
Current methods for handling recycled plastics primarily include burial, incineration, reutilization, and plastic oilification [
7,
8]. Burial is the most commonly used method for disposing of waste plastics, with approximately 14 million tons of used plastics being buried annually in China. However, waste plastics are difficult to degrade in natural environments. Plasticizers in the plastic material are hard to break down, leading to environmental Pollution of soil and water bodies caused by leakage of agents and additives; the incineration method can recover a large amount of heat, and the volume of waste plastic after incineration reduces by more than 90%. However, in addition to CO
2 and water, the byproducts of incineration also include harmful substances such as polycyclic aromatic hydrocarbons and acidic compounds, causing serious air pollution; the recycling method uses mechanical processing to handle plastic waste, converting it back into similar plastic products for reuse. But simple recycling results in low-quality plastic products, and mechanical processing requires substantial energy and human resources, with complex recycling processes. Pyrolysis of plastics through heating causes the breaking of C-C bonds within the plastic, accompanied by the breaking of C-H bonds, generating free radicals that continue to form various small-molecule hydrocarbon substances through different combinations of reactions, providing basic raw materials for various chemical products. However, pyrolysis processing has high requirements for the cleanliness, uniform quality, and chemical reagents of plastic waste, while in actual recycling processes, various types of plastics are often mixed together, making it difficult to unify the types of waste plastic raw materials from batch to batch. This poses high demands on pyrolysis technology, catalyst practicality, activity, and stability. In summary, the use of these physical and chemical methods for plastic recycling generally results in either low efficiency or follows a down-cycling route, where the recycled material is of lower quality. This leads to poor economic viability and significant secondary environmental pollution issues.
In recent years, the exploration of biodegradation and recycling technologies for plastic waste has become a hot topic both domestically and internationally [
9,
10]. By leveraging synthetic biology concepts, the construction of microbial or enzymatic elements with bio-polymerase functions can break down plastics into monomers under mild conditions without causing secondary pollution. Further, utilizing synthetic biology’s technical strategies to build biotransformation pathways from plastic degradation products to high-value commodities can achieve the “upcycling” of discarded plastic resources. Consequently, an integrated biological recycling process based on synthetic biology technology, which comprises “biodegradation, bioprocessing, and high-value biotransformation” is expected to be a significant approach for the recovery and disposal of plastic waste. This article reviews the latest research advancements in the screening of plastic-degrading microorganisms, the discovery and design of degradative enzymes, the elucidation of monomer degradation pathways, and the high-value biorefining of plastic degradation products, aiming to provide new perspectives for establishing more efficient.
2. Research Progress on Plastic Biodegradation
Plastics can be mainly classified into the following types based on their molecular composition and structure: polyolefins (PE, polypropylene (PP)), PVC, polystyrene (PS), PET, polyurethane (PU), polylactic acid (PLA), and polyhydroxyalkanoates (PHA), among others. Based on different depolymerization mechanisms, they can be further divided into hydrolytic plastics and non-hydrolytic plastics.
2.1. Screening of Microorganisms for Depolymerization of Hydrolytic Plastics and Mining of Depolymerizing Enzymes
Hydrolyzed plastics primarily refer to polymer plastics formed by the polymerization of monomers through ester bonds, with PET and PU as representative examples. The degradation of these polymers mainly occurs through the cleavage of ester bonds within the structure. Their biodegradation process is relatively simple. Currently, a variety of microorganisms from different bacterial genera have been identified as capable of degrading these types of plastics, and the degradation pathways are relatively well understood ().
PET plastic is a polymer compound formed by the ester bond linkage of terephthalic acid (TPA) and ethylene glycol (EG), typically existing in amorphous and semi-crystalline forms. It is mainly used in packaging materials such as beverage bottles [11]. Microorganisms degrade PET plastic by secreting extracellular depolymerases that hydrolyze the ester bonds in PET, leading to depolymerization. The resulting small molecular depolymerized products can then be further mineralized by the microorganisms into water and CO2 [12].
Crystallinity is one of the key factors influencing the biological depolymerization of PET. The higher the crystallinity, the more difficult the depolymerization becomes. Therefore, researchers typically use amorphous or low-crystallinity PET films as model substrates for screening PET-degrading microorganisms and studying their depolymerization characteristics. Reported PET-degrading microorganisms include Fusarium solani [13,14], Humicola insolens [15], Thermomonospora fusca [16,17,18,19], Streptomyces viridis [20], Brucella intermedia IITR130 [21], among others. However, most of these microorganisms only degrade and modify the surface of PET, and their ability to degrade actual PET plastic waste is quite limited. In 2016, Yoshida et al. discovered a bacterium, I. sakaiensis 201-F6, which was capable of fully degrading low-crystallinity PET films after 6 weeks at 30 °C. This bacterium is currently regarded as the most efficient in PET degradation among those reported to date [22]. Recently, Gordonia sp. CN2K has been shown to grow using PET as the sole carbon source, and it can degrade 40.43% of PET film within a 45-day incubation period [23].
Due to the high glass transition temperature of PET plastic, directly using enzymes to depolymerize PET at high temperatures (60–70 °C) has become a research hotspot in recent years. Various types of enzyme components with PET depolymerization activity have been discovered, including lipases, esterases, and keratinases, with keratinase being one of the most studied and efficient PET depolymerizing enzymes [24] (). Keratinase TfH from T. fusca DSM43793 causes a 50% loss in the mass of a PET film with a crystallinity of 10% under catalytic conditions at 55 °C within 3 weeks [25]. Keratinase HiC from Humicola insolens almost completely degrades low-crystallinity PET films under conditions of 70 °C within 96 h. It is currently the most active and thermally stable fungal-derived polyester hydrolase reported [26]. Keratinase LCC, derived from plant compost, can degrade 25% of amorphous PET films after 24 h of catalysis at 70 °C. This enzyme shows a certain degree of homology to TfH polyester hydrolase [27]. PETase from Deinococcus maricopensis was found to be comparable to LCCICCG at 50 °C in degrading semi-crystalline sections of post-consumer PET bottles, but it appeared to be less sensitive to crystallinity degree increase [28]. Additionally, lipases from Thermomyces insolens, Candida antarctica, and Aspergillus sp., as well as esterases from Cladosporium and Cladosporioides, Melanocarpus albomyces, and Penicillium citrinum, have some depolymerization effects on PET. However, these enzymes mainly increase the hydrophilicity of the PET surface and cause changes in its surface morphology, without significantly degrading the material.
PU plastic is a polymer formed by the condensation of three components: isocyanates, polyols, and chain extenders, which contain repeating urethane bonds. PU is a semi-crystalline thermosetting plastic, where isocyanates form its crystalline portion, known as the hard segment of PU, which determines the hardness and tensile strength of the material. Polyols and chain extenders make up the amorphous portion, known as the soft segment of PU, which governs the elasticity and elongation properties of PU. The biodegradation process of PU primarily occurs through the cleavage of ester bonds in the soft segments, leading to depolymerization. As a result, most microorganisms reported to degrade PU are focused on the degradation of polyester-based PU, with fewer reports on the degradation of polyether-based PU [29]. Several microorganisms, such as Fusarium sp., Curvularia sp., Cladosporium sp., Penicillium sp., Bacillus sp., Pseudomonas sp., and Streptomyces sp., have been confirmed to possess the ability to degrade PU plastics, as shown in . Álvarez-Barragán et al. [30] isolated six Cladosporium sp. strains, which were able to degrade 75–85% of water-based PU (Impranil DLN) within two weeks, showing a high degradation level for polyester-based PU. Aspergillus sp. is a type of fungus reported to degrade polyester-based PU. Aspergillus flavus, isolated from landfill soil, could use polyester PU films as the sole carbon source and degrade 60.6% of the polyester PU within 30 days [31]. Another Aspergillus sp. S45, isolated from a solid waste disposal site, could degrade 20% of polyester PU films in 28 days [32]. Aspergillus tubingensis, isolated from a landfill by Khan et al. [33], was able to degrade polyester PU films into fragments after 21 days of incubation in an inorganic salt liquid medium containing 2% glucose. Additionally, the microbial strain isolated from soil responsible for the maximum degradation of 55% of polyurethane films was identified as Aspergillus versicolor (ARF5) [34]. Streptomyces sp. PU10, isolated from soil contaminated with polyurethane, demonstrated the remarkable ability to degrade a soluble polyester-PU (Impranil) at various temperatures, achieving more than 96% degradation of 10 g/L within 48 h [35]. However, research on the use of these microorganisms to degrade real-world polyester PU plastic waste is still limited [36]. Nearly, a 42.1% weight loss of PU foam was observed after 30 days of exposure to Bacillus sp. YXP1, a bacterium isolated from a plastic landfill. This indicates that strain YXP1 exhibits exceptional degradation capabilities for PU foam [37].
.
Microorganisms responsible for depolymerizing plastics through hydrolysis.
| Plastic Classification |
Degradation Bacterias |
Degradation of Substrates |
Degradation Temperature (°C) |
Degradation Effect |
References |
| PET |
Fusarium solani |
PET fibers |
30 |
Surface modification of PET fibers |
[13,14] |
| Humicola insolens |
PET fibers |
30 |
Surface modification of PET fibers |
[15] |
| Thermobifida fusca |
PET fibers |
30 |
Surface modification of PET fibers |
[16,17,18,19] |
| Saccharomonospora viridis |
PET fibers |
30 |
Surface modification of PET fibers |
[20] |
| Ideonella sakaiensis 201-F6 |
PET film |
30 |
Complete degradation of low crystallinity PET film within 6 weeks |
[22] |
| Gordonia sp. CN2K |
PET sheet |
30 |
Degradation of 40.43% PET sheet in 45 days |
[23] |
| PU |
Aspergillus flavus
(ITCC 6051)
|
PU film |
28 ± 2 |
Degradation of 60.6% polyester PU film within 30 days |
[31] |
| Aspergillus tubingensis |
PU film |
37 |
Degradation of polyester PU film into fragments in 14 days |
[33] |
| Aspergillus sp. strain S45 |
PU film |
37 |
Degradation of 20% polyester PU film in 28 days |
[32] |
Cladosporium pseudocladosporioides, Cladosporium tenuissimum,
Cladosporium asperulatum,
Cladosporium montecillanum,
Aspergillus fumigatus,
Penicillium chrysogenum
|
PU film |
25–30 |
Degradation of 10% to 65% polyester PU film in 21 days |
[30] |
| Aspergillus versicolor |
PU film |
35 |
Degradation of 55% polyester PU film in 7 days |
[34] |
| Bacillus sp. YXP1 |
PU foam |
37 |
Degradation of 42.1% polyester PU foam in 30 days |
[37] |
Biological enzymes can depolymerize PU plastics by hydrolyzing the ester or urethane bonds. Common enzyme types include esterases, ureases, and proteases [38]. Esterases are currently the most effective enzymes for PU plastic degradation. For example, the esterase PudA is derived from Comamonas acidovorans TB-35 [39,40], and esterases PueA, PueB, and PulA from Pseudomonas species [41,42]. Although these cloned esterases can effectively depolymerize water-based polyurethane (Impranil DLN), they are largely ineffective against real PU waste. Enzyme (MTL) isolated from the gut bacteria of mealworms that consume plastic has the ability to hydrolyze thermoplastic films (PEGA-HDI) and thermosetting foams. It has been shown to possess superior PU plastic degradation capabilities compared to PueA, PueB, and PulA [43]. Papain, a protease, was able to break down PU films after being treated at 37 °C for 1 to 6 months. GPC and FTIR analysis showed some degree of carbamate bond cleavage [44]. Similarly, α-chymotrypsin could reduce the average molecular weight of PU by over 30% after 10 days of reaction at 25 °C [45]. Research on urease-mediated PU degradation is relatively scarce. Phua et al. [44] found that urease (EC 3.5.1.5) could degrade PU, with analysis via GPC and infrared showing breakdown primarily due to the hydrolysis of urethane bonds in PU. Yannick Branson and others discovered three urethanases (UMG-SP-1 to UMG-SP-3, GenBank accession codes: OP972509, OP972510, and OP972511). These urethanases were found from a soil-derived metagenomic library and were shown to effectively hydrolyze the urethane bonds in polyether-based polyurethanes [46]. Additionally, Schmidt et al. [47] discovered that keratinases LCC, TfCut2, Tcur1278, and Tcur0390, which degrade PET plastics, also exhibited some degradation of polyester-based PU. After 100 h of degradation at 70 °C, the PU plastic degradation rate reached 0.3–3.2%. Recently, cutinase PlCut1 [48] and CpCut1 [49] have demonstrated a strong ability to degrade PU, with CpCut1 showing particularly impressive results. It was capable of degrading 40.5% of thermoplastic PU film and 20.6% of post-consumer foam, while completely depolymerizing Impranil DLN-SD.
.
Depolymerases responsible for depolymerizing plastics through hydrolysis.
| Plastic Classification |
Degradation of Substrates |
Source of Depolymerase |
Depolymerase |
Degradation Temperature (°C) |
Degradation Effect |
References |
| PET |
Beverage bottle |
Thermobifida fusca DSM43793 |
TfH |
55 |
Quality loss of 50% in 21 days |
[25] |
| Low crystallinity (7%) PET film |
Humicola insolens |
HiC |
70 |
Quality loss of 97% in 96 h |
[26] |
| PET film |
Plant compost |
LCC |
70 |
Quality loss of 25% in 24 h |
[27] |
| Low crystallinity (1.9%) PET film |
Ideonella sakaiensis 201-F6 |
PETase |
30 |
— |
[27] |
| Semi-crystalline sections of PET bottles |
Deinococcus maricopensis |
DmPETase |
50 |
— |
[28] |
| PU |
Impranil DLN |
Aspergillus flavus
(ITCC 6051)
|
PudA |
45 |
— |
[39,40] |
| Impranil DLN |
Aspergillus tubingensis |
PulA |
48 |
— |
[50] |
| Impranil DLN |
Aspergillus sp. S45 |
PueA/PueB |
65/60 |
— |
[42,43] |
| Polyester PU |
T. Fusca KW3 |
TfCut2 |
70 |
Quality loss of 1.9% in 100 h |
[48] |
| Polyester PU |
Plant compost |
LCC |
70 |
Quality loss of 3.2% in 100 h |
[48] |
| PU foam |
Gut bacterium of mealworms |
MTL |
70 |
— |
[44] |
| PU foam |
— |
Urethanases |
70 |
— |
[47] |
2.2. Screening of Non-Hydrolytic Plastic-Degrading Microorganisms and Exploration of Depolymerizing Enzymes
Non-hydrolytic plastics refer to high molecular weight plastics made from olefin monomers, represented by PE and PS. The chemical structure of their main chains consists of alkyl carbons, and the C-C bonds are highly inert with a high reaction energy barrier. This makes them difficult to break down, which is a key reason why these types of plastics are challenging for microorganisms to degrade [
51].
Researchers have isolated several microorganisms capable of degrading PE-type plastics, as shown in . Qiuxia Han et al. [
52] isolated
Aspergillus niger M6 from farmland soil, which could use modified PE film as its sole carbon source, achieving a 20% mass loss of PE film after 30 days. Balasubramanian et al. [
53] isolated 15 high-density PE-degrading bacteria from a plastic waste dump site in Manaw Bay, India. Among them,
Arthrobacter sp. GMB5 and
Pseudomonas sp. GMB7 degraded 12% and 15% of PE film, respectively, in 30 days. Tribedi et al. [
54] enriched and isolated a
Pseudomonas sp. AKS2 strain capable of degrading low-density PE from soil, which caused a 4–6% mass loss of PE in 45 days. Similarly,
Rhodococcus sp. C208 was enriched from abandoned agricultural plastic film and achieved a degradation rate of 0.86% per week [
55]. It has been confirmed that
Klebsiella pneumoniae CH001 efficiently degraded high-density PE by forming biofilms and altering surface properties. Universal Tensile Machine analysis revealed a significant decrease in weight (18.4%) and a reduction in tensile strength (60%) of high-density PE film [
56].
Bacillus sp. strain PE3 produces ligninolytic enzymes (laccase, lignin peroxidase) and lipopeptide biosurfactants in media with PE as a carbon source, reflecting its ability to degrade PE [
57].
P. aeruginosa V1 demonstrated the highest CO
2 evolution (8.86 g/L) during LDPE degradation, confirming PE biodegradation [
58]. The degradation of high-density PE, and linear low density PE supported by Fourier transform infrared spectrometer, mass loss, scanning electron microscope, X-ray diffraction, X-ray photoelectron spectroscopy, and gas chromatography-mass spectrometry analyses, confirming the strain
Enterobacter hormaechei had the ability to degrade multiple PE [
59].
There are fewer reports on microorganisms that degrade PS plastics. Eisaku et al. [
60] first isolated five PS-degrading microorganisms from soil, including
Xanthomonas sp.,
Sphingobacterium sp., and
Bacillus sp. Additionally, Ji Rong et al. [
61] applied
Penicillium variabile CCF3219 to 14C-labeled PS films pretreated with ozone oxidation, achieving nearly complete mineralization to CO
2 and water over 16 weeks. In recent years, the use of insect gut microbiota for degrading polyolefin plastics has developed rapidly. Yang Jun et al. [
62,
63] discovered that the larvae of
Plodia interpunctella Hübner (Indian meal moth) could feed on PE films, and two PE-degrading bacteria were isolated from their gut:
Enterobacter asburiae YT1 and
Bacillus sp. YP1. The same team also found that the larvae of
Tenebrio molitor Linnaeus (yellow mealworm) exhibited some degradation ability for PS films and further isolated a PS-degrading bacterium,
Exiguobacterium sp. YT2, which degraded 7.4% of PS within 60 days [
64]. The degradation rate of polystyrene (PS) by marine microorganisms Gordonia sp. and Novoshinegobium sp. at the laboratory level over one month ranged from 2.7% to 7.7% [
65]. Eight isolated species from mealworms (
Acinetobacter septicus,
Agrobacterium tumefaciens,
Klebsiella grimontii,
Pseudomonas multiresinivorans,
Pseudomonas nitroreducens,
Pseudomonas plecoglossicida,
Serratia marcescens, and
Yokenella regensburgei) were identified that degrade PS [
66]. The gut microbiota of
Zophobas atratus larvae has been shown to have the ability to degrade PS under both anaerobic and aerobic conditions, with anaerobic conditions favouring a more active plastic degradation [
67]. In future studies, insect gut microbiota will provide an important source for screening microorganisms capable of efficiently degrading polyolefin plastics.
.
Microorganisms responsible for depolymerizing plastics through non-hydrolysis.
| Plastic Classification |
Degradation of Substrates |
Degradation Bacterias |
Degradation Temperature (°C) |
Degradation Effect |
References |
| PE |
Modified PE film |
Aspergillus niger M6 |
28 |
Quality loss of 20% in 30 days |
[52] |
| High density PE |
Arthrobacter sp. GMB5 |
30 |
Quality loss of 12% in 30 days |
[53] |
| High density PE |
Pseudomonas sp. GMB7 |
30 |
Quality loss of 15% in 30 days |
[53] |
| High density PE |
Klebsiella pneumoniae CH001 |
30 |
Quality loss of 18.4% in 60 days |
[56] |
| Low density PE film |
Enterobacter asburiae YT1 |
30 |
Quality loss of 6.1% ± 0.3% after 60 days |
[54] |
| Low density PE film |
Bacillus sp. YP1 |
30 |
60 days quality loss 10.7% ± 0.2% |
[54] |
| PS |
PS foam |
Mealworms
(the larvae of Tenebrio molitor Linnaeus)
|
30 |
30 days quality loss of 31.0% ± 1.7% |
[64] |
| PS film |
Exiguobacterium sp. YT2 |
30 |
Quality loss of 7.4% ± 0.4% after 60 days |
[64] |
| PS film |
Penicillium variabile |
24 |
Mineralization can be completed
within 16 weeks |
[61] |
Many types of enzymes participate in the biodegradation process of non-hydrolyzable plastics such as PE and PS. Enzymes like laccase, manganese peroxidase (MnP), lignin peroxidase (LiP), and alkane hydroxylases (AH) have been shown to have certain degradation effects on pre-treated non-hydrolyzable plastics [
68]. In the presence of Cu
2+, when PE plastic is treated with laccase derived from
Rhodococcus ruber C208, a significant increase in carbonyl content is observed, and the weight-average molecular weight (
Mw) and number-average molecular weight (
Mn) of the polymer decrease by 20% and 15%, respectively. The oxidation and cleavage occur primarily in the amorphous regions of the PE film [
69]. Laccase from
Trametes versicolor, in the presence of 1-hydroxybenzotriazole, accelerates the degradation rate of PE films [
70]. The alkane hydroxylase family, including AH, can degrade carbon-hydrogen compounds through oxidation at the terminal or sub-terminal positions [
56]. In
Escherichia coli BL21, the gene encoding AH from
Pseudomonas sp. E4 was expressed exogenously, and after 80 days of cultivation at 37 °C, 20% of low molecular weight PE was converted to CO
2 [
71]. Further integration of the AH catalytic system from
Pseudomonas aeruginosa E7 (including alkane monooxygenase, redox enzymes, and reductase) into engineered
Escherichia coli BL21 improved the PE degradation efficiency to 30% [
72]. Currently, reports on PS depolymerase are limited, with one notable example being a non-heme hydroquinone peroxidase from the lignin-decolorizing bacterium
Azotobacter beijerinckii HM121. This enzyme can degrade insoluble PS in a biphasic system (dichloromethane-water). In the presence of hydrogen peroxide and tetramethylhydroquinone, PS can be degraded into water-soluble small molecular products within 5 min [
73] ().
.
Depolymerases responsible for depolymerizing plastics through non-hydrolysis.
| Plastic Classification |
Degradation of Substrates |
Source of Depolymerase |
Depolymerase |
Degradation Temperature (°C) |
Degradation Effect |
References |
| PE |
Oxidized low-density PE |
Phanerochaete chrysosporium MTCC-787 |
LiP/MnP |
37 |
Degradation of 70% within 15 days |
[68] |
| Low molecular weight PE powder |
Pseudomonas sp. E4 |
alkB |
37 |
Degradation of 20% within 80 days |
[71] |
| Low molecular weight PE powder |
Pseudomonas aeruginosa E7 |
AH system |
37 |
Degradation of 30% within 80 days |
[72] |
| PS |
PS |
Azotobacter beijerinckii
HM121
|
Non-heme hydroquinone
Peroxidase
|
30 |
Degradation of PS within 5 min |
[73] |
3. Design and Modification of Plastic Depolymerase
At present, the plastic depolymerase gene library has problems such as low catalytic efficiency, poor stability, and low expression levels, which limit the large-scale production and application of plastic depolymerase. By utilizing protein engineering techniques such as rational design and targeted modification, it is expected to improve the activity, stability, and specificity of plastic depolymerase.
3.1. Improving the Thermal Stability of Enzymes
The protein engineering technology method is expected to improve the activity, stability, and specificity of plastic depolymerase.
The glass transition temperature is one of the important factors affecting the biodegradation of plastics. At the glass transition temperature, the amorphous part of PET plastic has higher flexibility, making it easier for enzymes to contact and depolymerize. The glass transition temperature of PET is around 65 °C, so it is required that the depolymerase has strong heat resistance [
74]. Divalent metal ions (Ca
2+and Mg
2+) can increase the melting point temperature of the polyester hydrolase TfCut2 by 10.8–14.1 °C, thereby improving its thermal stability. The modified enzyme can improve the hydrolysis efficiency of semi crystalline PET to 12.9% under 65 °C and 48 h treatment conditions [
19]. By introducing disulfide bonds into TfCut2, its melting point temperature was increased by 25 °C and its half deactivation temperature was increased by 17 °C. The enzyme mutant was able to depolymerize 25% PET film under 70 °C and 48 h treatment conditions, and its catalytic efficiency was doubled compared to the original enzyme [
18]. By introducing disulfide bonds into metal-binding sites of Thc_Cut1, the variant exhibited high enzymatic activity at elevated temperatures (70 °C). After 96 h of hydrolysis using the variant, 96.2% of post-consumer PET bottle particles (without energy-intensive molten quenching pretreatment) were successfully degraded, which is 87.5 times higher than using the wild-type Thc_Cut1 [
75]. In addition, the Marty team from the University of Toulouse in France [
76] increased the melting point temperature of LCC by 9.8 °C by introducing disulfide bonds to replace amino acid residues near the binding site of LCC metal ions, thereby improving its degradation efficiency for PET plastics. Therefore, enhancing the thermal stability of enzymes can effectively improve their catalytic efficiency towards plastic substrates.
3.2. Increasing the Substrate Binding Pocket of Enzymes
One of the most commonly used strategies to enhance enzyme catalytic efficiency is to modify the catalytic center of enzymes to promote their binding with substrates [
77]. If the substrate binding pocket can be expanded to make it easier for plastic macromolecules to enter the catalytic center of enzymes, it will enhance the catalytic activity of plastic degrading enzymes. Araújo et al. [
13] mutated the amino acids near the active center of keratinase FsC and expanded the substrate binding pocket using the alanine substitution method. The mutant L182A and L81A showed a 4-fold and 5-fold increase in hydrolysis activity of PET fibers, respectively; the Marty team from the University of Toulouse in France [
76] identified the key amino acid residues that affect enzyme catalytic efficiency by regulating the substrate binding pocket of LCC and the coincidence of PET substrate. They changed all amino acid residues in the catalytic groove by arranging and combining them. Among them, the “grooves” on the surfaces of F243I and F243W were the easiest to fit with PET, and the mutant significantly improved enzyme catalytic efficiency. On this basis, disulfide bonds were further introduced to replace amino acid residues near the metal ion binding site to improve the thermal stability of the enzyme. The resulting enzyme mutant can depolymerize over 90% of bottle grade PET within 10 h.
3.3. Enhancing Enzyme and Substrate Accessibility
The hydrophobic properties of polymer surfaces prevent the hydrophilic groups on the surface of depolymerase from effectively adsorbing onto the polymer surface, thereby reducing the accessibility of the enzyme to the substrate. If a hydrophobic binding module can be introduced into PET depolymerase, it will increase the accessibility of the enzyme to the substrate. Ribitsch et al. [
78] fused PET keratinase Thc_Cut1 with two hydrophobic proteins from
Trichoderma, HFB4 and HFB7, and the resulting fusion enzyme increased the hydrolysis efficiency of PET by more than 16 times. Ribitsch et al. [
79] increased the hydrolytic activity of PET by 3.8 times by introducing two hydrophobic proteases into the PET keratinase (Thc_Cut1): the cellulase (CBM) from
Hypocrea jecorina and the polyhydroxyalkanoate depolymerase (PBM) from
Alcaligenes faecalis. Similarly, Gamerith et al. [
80] introduced the PBM from
A. faecalis into the polyamidease PA of
Nocardia farcinica, and the fused enzyme PA_PBM exhibited three times higher hydrolytic activity on the polymer compared to PA. Therefore, increasing the accessibility of enzymes and substrates is an effective strategy to enhance the hydrolytic activity of depolymerases towards hydrophobic polymers. In addition, increasing the hydrophobicity of enzyme molecule substrate binding pockets and promoting the adsorption and binding of hydrophobic plastic polymers with enzymes to improve enzyme catalytic efficiency is another method to enhance enzyme substrate accessibility. For example, Silva et al. [
81] increased the hydrophobicity of the substrate binding pocket of the keratinase Tfu_0883 by designing a double mutant Q132A/T101A. The mutant showed a 1.6-fold increase in PET fiber degradation activity. The last few years have witnessed impressive progress in tailoring natural enzymes by computational redesign strategies. Qi Qingsheng et al. conducted kinetic analysis of substrate enzyme complexes by introducing dynamic protein conformation information of PET degrading enzymes. They developed LCC mutants with significantly increased affinity for PET substrates, which degraded over 90% of PET within 3.3 h [
82]. Bian Wu et al. employed a hybrid computational strategy to redesign a PET hydrolase named TurboPETase that significantly outperforms other well-known PET hydrolases. Nearly full degradation of postconsumer PET bottles was achieved at an industrially relevant level of solids loading. Kinetic and structural analysis suggests that the improved performance may be attributed to a more flexible PET-binding groove that facilitates the targeting of more specific attack sites [
83]. Molecular dynamics simulations, which provided a thorough analysis of the UMG-SP-2 structure, demonstrated that the A141G mutation leads to a significantly stronger binding of the substrate compared to the wild-type (WT) enzyme. Over the course of 1.2 µs of simulations, the A141G mutant formed, on average, 36 more protein-substrate contacts than the WT system. Additionally, the root-mean-square deviation (RMSD) of the MDI-DEG substrate atoms in the A141G mutant was 0.6 Å lower than that in the WT enzyme, indicating a higher level of substrate stability [
84].
3.4. Reduce Enzyme Product Inhibition
After hydrolysis of PET, degradation monomers such as bis(hydroxyethyl) terephthalate (BHET), mono(hydroxyethyl) terephthalate (MHET), TPA, and EG are produced. Among these, BHET and MHET competitively bind to the substrate-binding site of PET hydrolytic enzymes, thereby inhibiting the depolymerization activity of PET enzymes. To address this issue, Wei et al. [
85] mutated the amino acid residue at position 62 of TfCut2, and the resulting G62A mutant no longer interacted with MHET. The binding constant of this enzyme to MHET decreased to 2/11 of the original, and the PET degradation efficiency increased by 2.7 times. Carniel et al. [
86] combined the lipase CalB from
C. antarctica with HiC to eliminate the accumulation of MHET, a degradation product of PET. The dual-enzyme system exhibited a 7.7-fold increase in PET degradation efficiency compared to the single-enzyme catalytic system. Barth et al. [
87] developed an immobilized dual-enzyme system (TfCa-TfCut2 and TfCa-LCC) for PET degradation, where TfCa was used to hydrolyze intermediate products BHET and MHET. Compared to single-enzyme treatments, the synergistic effect of the dual-enzyme system increased the production of BHET and MHET by 91% and 104%, respectively, thereby improving PET degradation efficiency. A dual-enzyme system consisting of FastPETase and a redesigned thermophilic carboxylesterase (KL-MHETase) can effectively eliminate the accumulation of the intermediate MHET. This system demonstrates a PET depolymerization efficiency 2.6 times faster than FastPETase alone. The synergistic effect enhances the yield of TPA by 1.75 times and increases the purity of TPA to 99.5% [
88].
4. The High-Value Biorefining of Plastic Degradation Products
Establishing a bioconversion technology system from plastic degradation products to valuable chemicals can not only promote the development of the circular economy, but also effectively conserve non-renewable resources such as petroleum and natural gas, reduce greenhouse gas emissions, and protect the ecological environment. However, waste plastics are complex in structure, difficult to classify, and have diverse degradation products. To achieve high-value biorefining of plastic degradation products, both chemical and biological methods are required for the depolymerization process. Whether the plastic monomers are obtained through biological or chemical strategies, after identifying the small molecules or oligomers resulting from degradation, microorganisms that can utilize these small molecules should be selectively explored. Their degradation pathways should be analyzed, and synthetic biology techniques can be employed to design and construct pathways for converting plastic degradation products into high-value chemicals. This approach holds promise for establishing a “plastics recycling” circular economy for waste plastic resources.
4.1. Biological Utilization Pathways of Plastic Degradation Products
The complexity of structure and differences in depolymerization conditions lead to a diverse composition of plastic degradation products, mainly including organic acids, alcohols, aromatic compounds, and fatty hydrocarbons. A large body of research has been conducted by domestic and international researchers on the biological utilization of these plastic monomers, achieving significant progress in various stages.
4.1.1. Organic Acid Plastic Degradation Products
Adipic Acid
Adipic acid, one of the degradation products of plastics such as PU, has its metabolic pathway elucidated in Acinetobacter [
89]. As shown in , adipic acid first forms adipoyl-CoA under the catalysis of succinyl-CoA transferase (DcaIJ), and then, under the catalysis of acyl-CoA dehydrogenase (DcaA), it generates 5-carboxy-2-pentenoil-CoA. This is further converted into 3-hydroxyadipoyl-CoA by the action of enoyl-CoA hydratase (DcaE), followed by its conversion to 3-ketoadipoyl-CoA by the action of 3-hydroxyadipoyl-CoA dehydrogenase (DcaH). Finally, 3-ketoadipoyl-CoA is converted into succinyl-CoA and acetyl-CoA through the action of acyl-CoA thioesterase (DcaF), which then enters the TCA cycle for cell growth and metabolism.
. Biological degradation pathway for plastics from organic acid based monomers (adipic acid, 6-hydroxyhexanoic acid). (Key enzymes in metabolic pathway: DcaIJ—succinyl-CoA transferase; DcaA—acyl-CoA dehydrogenase; DcaE—enoyl-CoA hydratase; DcaH—3-hyroxyacyl-CoA dehydrogenase; DcaF—acyl-CoA thiolase; ChnD—6-hydroxyhexanoate dehydrogenase; ChnE—6-oxohexanoate dehydrogenase).
6-Hydroxyhexanoic Acid
6-Hydroxyhexanoic acid, present in PU plastic degradation products, can be converted into adipic acid through a two-step process catalyzed by 6-hydroxyhexanoic acid dehydrogenase (ChnD) and 6-oxocaproic acid dehydrogenase (ChnE) within the microbial cell, as shown in . Subsequently, adipic acid follows the above-mentioned degradation pathway, ultimately forming succinyl-CoA and acetyl-CoA, which enter the TCA cycle [
90].
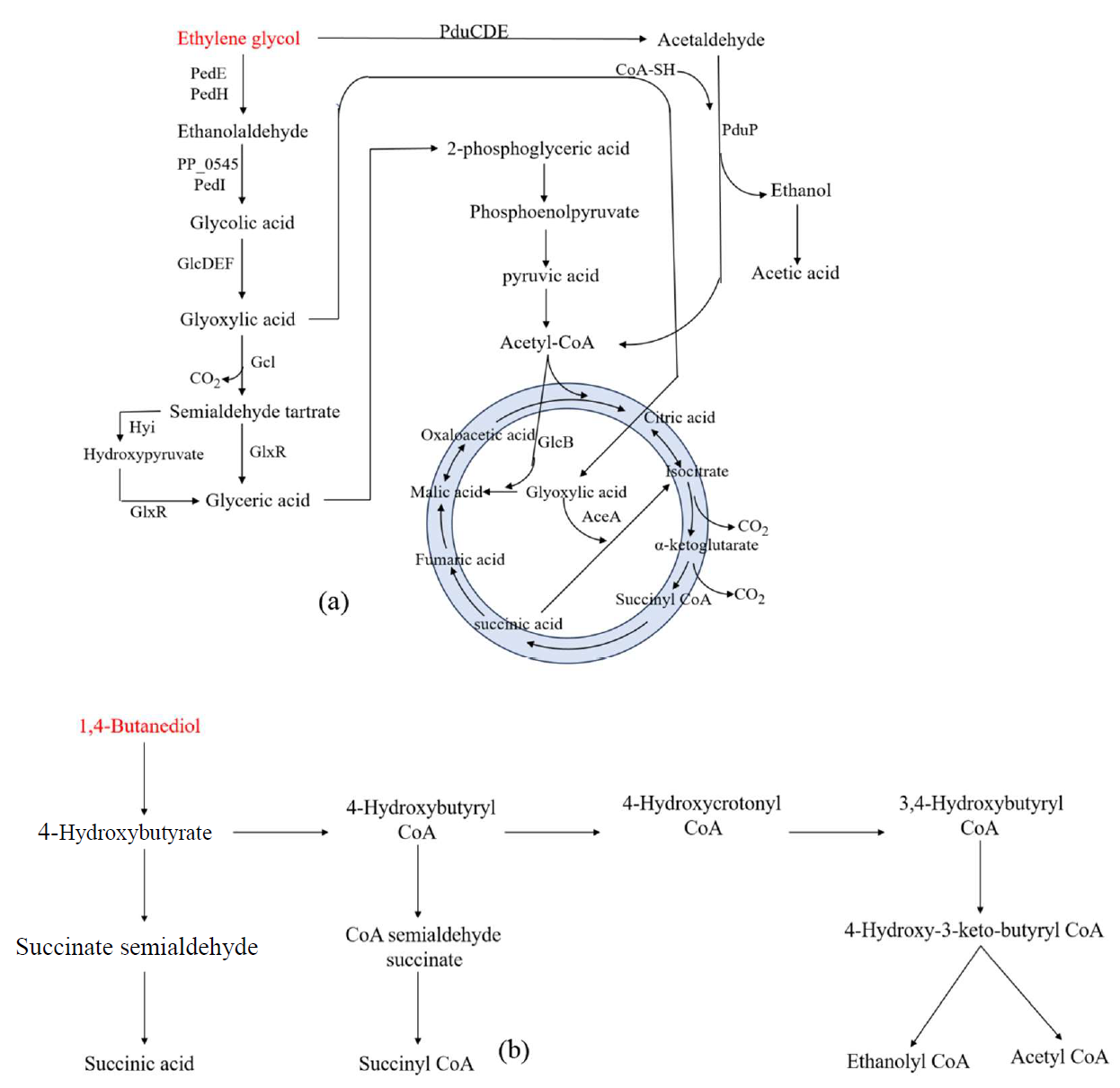
4.1.2. Degradation Products of Organic Alcohol Plastics
EG
EG is one of the depolymerization products of plastics such as PET and PU. In recent years, there has been widespread attention on the biotransformation of EG into higher-value biochemicals such as glycolic acid, glyoxylic acid, and rhamnolipid using soil microorganisms like
Pseudomonas putida (
P. putida) [
91]. As shown in a, in
P. putida KT2440, EG is first converted to glycolaldehyde under the catalysis of the periplasmic pyrroloquinoline quinone (PQQ)-dependent alcohol dehydrogenases (PedE, PedH). This is followed by the conversion to glycolic acid by the cytoplasmic aldehyde dehydrogenases PP_0545 and PedI, and subsequently, glycolic acid is converted to glyoxylic acid under the action of the membrane-bound glycolic acid oxidase GlcDEF. The generated glyoxylic acid can enter the TCA cycle through two pathways: (1) It condenses with succinate to form isocitrate under the catalysis of isocitrate lyase (AceA), and then enters the TCA cycle. (2) It condenses with acetyl-CoA to form malate under the catalysis of malate synthase (GlcB), entering the TCA cycle [
91]. However, in pathway (1), during the conversion of isocitrate to succinate, two molecules of CO
2 are released, which means the two carbon atoms provided by EG cannot contribute to the central metabolism to support cell growth. In pathway (2), the shortage of acetyl-CoA limits the further conversion of glyoxylic acid into malate, thus restricting its entry into central metabolism. Therefore,
P. putida KT2440 cannot use EG as the sole carbon source for growth [
91]. On the other hand,
P. putida JM37 has another metabolic pathway for EG, where glyoxylic acid is carboxylated by glyoxylic acid carboxylase (Gcl) to form tartronate semialdehyde, which is then converted to glycerate by hydroxypropionate isomerase (Hyi) and tartronate semialdehyde reductase (GlxR). The glycerate is further converted to 2-phosphoglycerate, which enters the glycolysis pathway and eventually the TCA cycle [
92]. Therefore,
P. putida JM37 can grow well on a medium with EG as the sole carbon source. Through a comprehensive investigation of the expression and transcription levels of various gene elements in the EG metabolism pathway, it was found that Gcl and GlxR are key nodes in the EG utilization process.
Odorous pseudomonas expressing both Gcl and GlxR achieved rapid growth on a medium with EG as the sole carbon source [
91]. In addition, in
Acetobacter woodii, Trifunović et al. [
93] found that the
pdu gene cluster encodes for propylene glycol dehydration (PduCDE) and CoA dependent acetaldehyde dehydrogenase (PduP), which are oxidized to acetic acid, and the resulting reduced equivalents are used to fix CO
2 and synthesize acetic acid in the Wood Ljungdahl pathway.
. Biological degradation pathway for plastic from organic alcohol based monomers: (<b>a</b>) ethylene glycol; (<b>b</b>) 1,4-butanediol. (Key enzymes in metabolic pathway: PedE, PedH—quinoprotein alcohol dehydrogenase; PP_0545 and PedI—aldehyde dehydrogenase; GlcDEF—glycolate oxidase; Gcl—glyoxylate carboligase; Hyi—hydroxypyruvate isomerase; GlxR—artronate semialdehyde reductase; AceA—isocitrate lyase; GlcB—malate synthase; PduP—propionaldehyde dehydrogenase).
1,4-Butanediol
1,4-Butanediol is one of the main degradation products of PU.
P. putida KT2440 can grow on a medium with 1,4-butanediol as the sole carbon source, although its growth rate is very slow. Through adaptive evolution, the efficiency of
P. putida KT2440 in utilizing 1,4-butanediol can be improved. Based on this, Li et al. [
94] conducted genomic sequencing and proteomic analysis of mutant strains to elucidate the biodegradation pathway of 1,4-butanediol. As shown in b, 1,4-butanediol is first oxidized to 4-hydroxybutyrate, a process primarily catalyzed by high-expressing dehydrogenases encoded by the
ped gene cluster (PP_2674–2680). The resulting 4-hydroxybutyrate can be metabolized through three pathways: (1) It is further oxidized to succinate by dehydrogenases encoded by the
ped gene cluster. (2) It is activated by acyl-CoA synthase AcsA1 (PP_4487) and converted to succinyl-CoA. (3) It undergoes β-oxidation to form ethanolyl-CoA and acetyl-CoA. Currently, only the third pathway has been confirmed, while the succinate synthesis and succinyl-CoA synthesis pathways remain to be further verified.
4.1.3. Aromatic Plastic Degradation Products
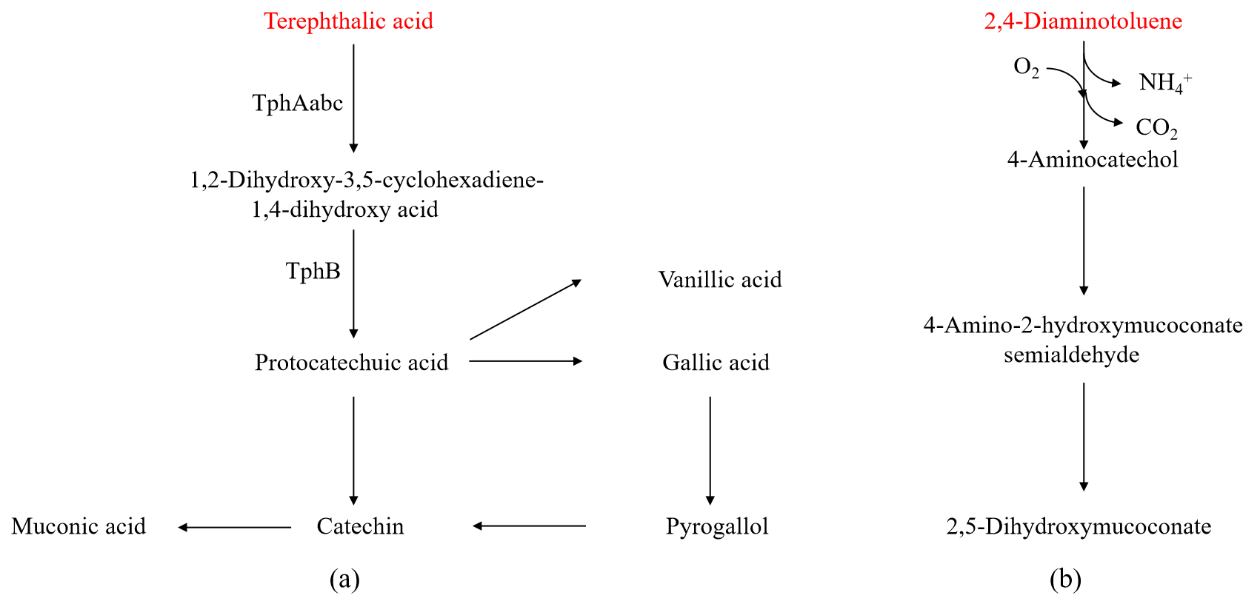
TPA
TPA is another important degradation product of PET. Converting TPA into higher-value aromatic compounds can improve the economic viability of recycling PET plastic products. Microorganisms such as
Comamonas sp. [
95],
Delftia tsuruhatensis [
96],
Rhodococcus sp. [
97], and
Rhodococcus erythropolis [
98] can grow and metabolize on media where TPA serves as the sole carbon source. As shown in a, the degradation pathway of TPA within the microbial cell involves two steps: TPA is converted into an important intermediate, protocatechuic acid, through the action of 1,2-dioxygenase (TphAabc) and 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase (TphB) [
99]. Protocatechuic acid is a simple phenolic acid that can be further converted into high-value aromatic compounds such as gallic acid, resorcinol, mucic acid, and vanillic acid, or used to synthesize important bioproducts such as rhamnolipids and PHA. The low water solubility and low fermentation substrate concentration of TPA are among the bottlenecks limiting its use as a fermentation substrate. To address this issue, Kenny et al. [
100] used a co-substrate of TPA and glycerol for the cultivation and fermentation of
P. putida GO16, achieving a PHA production rate of 108.8 mg (/L·h).
. Biological degradation pathway for plastics from aromatic monomers: (<b>a</b>) terephthalic acid; (<b>b</b>) 2,4-diaminotoluene. (Key enzymes in metabolic pathway: TphAabc-TPA 1,2-dioxygenase; TphB-1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase).
2,4-Diaminotoluene
A depolymerization product of plastics such as PU. In 2020, Espinosa et al. [
101] isolated a strain of
Pseudomonas TDA1 from plastic landfill soil, which can utilize PU degradation oligomers and 2,4-diaminotoluene as carbon/nitrogen sources for growth. Through genomic analysis, they proposed a preliminary degradation pathway for 2,4-diaminotoluene. As shown in b, the methyl group of 2,4-diaminotoluene undergoes oxidation, decarboxylation, and deamination to form 4-amino-resorcinol. 4-amino-resorcinol may be converted into 4-amino-2-hydroxy-mucic acid in the form of a diol and further degraded through a metabolic pathway similar to that of catecholic acid. In the future, comprehensive proteomic and transcriptomic analyses of the relevant genes in the predicted metabolic pathway are required, along with an investigation of the distribution and flux changes of metabolic intermediates, in order to accurately map the degradation pathway of 2,4-diaminotoluene.
4.1.4. Aliphatic Hydrocarbon Plastic Degradation Products
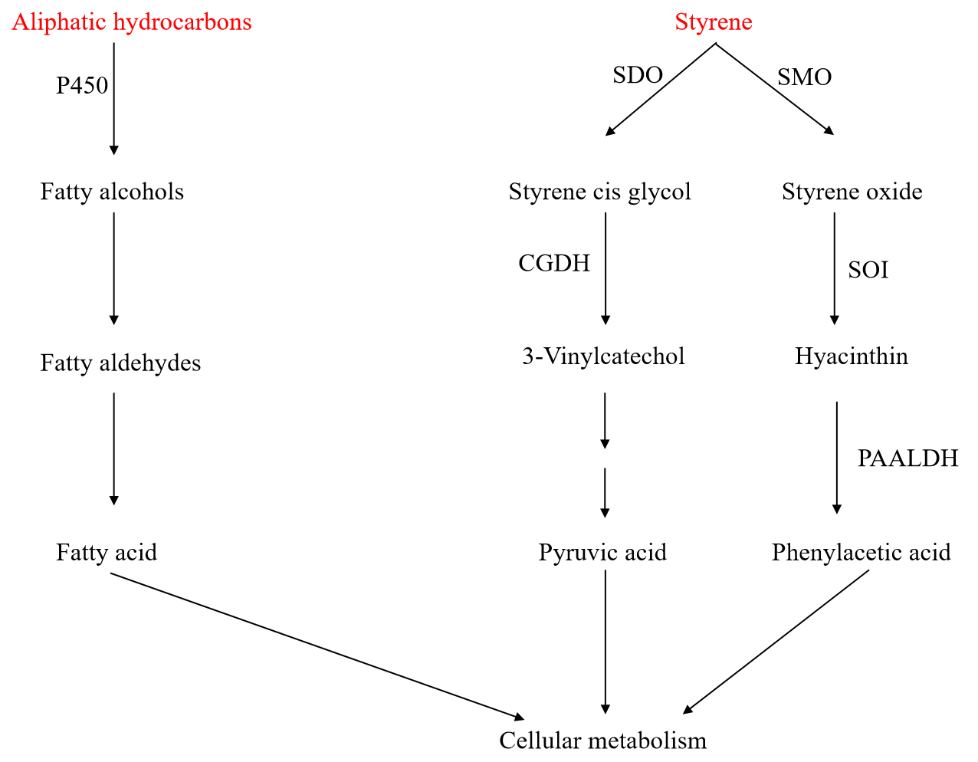
Various small aliphatic hydrocarbons can be obtained through the pyrolysis of plastic products such as PE, PP, and PVC. The metabolic pathways of aliphatic hydrocarbons are widely present in nature. In eukaryotic microorganisms such as
Candida sp. and
Yarrowia lipolytica, aliphatic hydrocarbons are first oxidized by the CYP52 family of cytochrome P450 monooxygenases in the endoplasmic reticulum to form fatty alcohols. The fatty alcohols are then further oxidized to fatty aldehydes in the endoplasmic reticulum or peroxisomes, and subsequently oxidized to fatty acids. Fatty acids are catalyzed by acyl-CoA to form acyl-CoA. Acyl-CoA can either be used for triglyceride synthesis or undergo complete oxidation through β-oxidation in peroxisomes to form acetyl-CoA, which enters central metabolic pathways [
102].
For benzene-based aliphatic hydrocarbons, their degradation pathways mainly include ring cleavage and side-chain oxidation, as shown in . Ring cleavage degradation has been reported infrequently and is primarily found in
Rhodococcus species [
103]. The aromatic ring of styrene is first hydroxylated by styrene dioxygenase (SDO) to form styrene cis-glycol, which is then further oxidized by cis-glycol dehydrogenase (CGDH) to form 3-vinylphenol, which is subsequently converted into pyruvate and enters central metabolism. The ring cleavage pathway is non-specific and can act on any substance containing a benzene ring structure. The oxidation of the vinyl side chain is the main degradation pathway for styrene and is widely present in microorganisms such as
Pseudomonas sp. [
104],
Corynebacterium sp. [
105], and
Rhodococcus sp. [
106]. Styrene undergoes transformation into phenylacetic acid through enzymatic actions of styrene monooxygenase (SMO), styrene oxide isomerase (SOI), and phenylacetaldehyde dehydrogenase (PAALDH). After further hydroxylation, phenylacetic acid undergoes β-oxidation to form acetyl-CoA, which enters the TCA cycle or is converted into PHA. Additionally, the styrene oxidation intermediate phenylacetic acid can also serve as an important precursor for the synthesis of high-value chemicals such as phenylethanol and phenylethylamine [
107].
. Biological degradation pathway for plastics from aliphatic hydrocarbon monomers (Key enzymes in metabolic pathway: P450—monooxygenase P450; SDO—styrene dioxygenase; SMO—styrene monooxygenase; CGDH—cis-ethylene glycol dehydrogenase; SOI—styrene oxide isomerase; PAALDH—phenylacetaldehyde dehydrogenase).
PHAs are intracellular carbon sources and energy reserves in most bacteria, and because they are completely biodegradable, they are considered a promising alternative to traditional plastics as a new type of biopolymer material. In recent years, the synthesis of PHA using plastic degradation products as substrates has received widespread attention. In 2011, Jasmina et al. [
108] utilized
P. putida CA-3 to convert styrene and achieved a PHA production of 3.36 g/L, establishing a unique connection between the degradation of aromatic environmental pollutants and the synthesis of aliphatic PHA, thus providing a feasible approach for the recycling of PS plastics. Further, using low-density PE powder as the substrate, after 21 days of bioconversion, Sen et al. [
109] reported the accumulation of short-chain PHA with a cell dry weight of 3.18% in
Cupriavidus necator H6. This was the first report on the direct degradation of PE materials and the subsequent synthesis of bio-based compounds. There are also reports of using the pyrolysis products of processed olefin plastics to cultivate bacteria for PHA production. Among them,
Pseudomonas aeruginosa PAO-1 uses thermally decomposed hydrocarbons as a carbon source for fermentation, which can produce PHA accounting for 25% of the cell dry weight [
110]. A platform has been developed for the conversion of short chain diols and long-chain dicarboxylic acids into PHA, which can convert diols such as 1,3-propanediol, 1,4-butanediol, and 1,5-pentanediol, as well as chain dicarboxylic acids such as adipic acid, into PHA, expanding the availability of these plastic degradation products [
111]. In addition, it was found that
P. putida GO16,
P. putida GO19, and
Pseudomonas frederiksbergensis GO23 can accumulate a certain amount of medium length PHA while degrading PET. The accumulation rate of PHA by GO16 and GO19 can reach 8.4 mg/(L·h) [
112].
Surfactants can emulsify hydrophobic substances in aqueous media, thereby increasing the utilization of hydrophobic substances by cells. Therefore, hydrophobic substrates such as hydrophobic fatty hydrocarbons obtained by pyrolysis of plastics such as PE, PP, PVC,
etc. are commonly used in the synthesis research of surfactants. For example, the Salmonella bacterium
Renibacterium salinarum 27BN can grow and accumulate rhamnose esters using n-hexadecane as the sole carbon source, and the secretion of rhamnose esters can further promote the utilization of hexadecane [
113]. It is worth mentioning that the synthesis of rhamnose esters shares the R-3-hydroxyalkanoic acid precursor library with PHA. Therefore, many microorganisms capable of assimilating plastic degradation products to synthesize PHA also have the potential to synthesize rhamnose esters [
114].
Oil is the main substance for energy in microorganisms, and oil producing microorganisms can convert fatty hydrocarbons and plastic degradation products to synthesize and accumulate oil.
Y. Lipolytica strain 78-003 can directly utilize PP plastic pyrolysis mixture (mainly containing fatty alcohols, alkanes, and alkenes), with a cell biomass of 2.34 g/L, an oil content of 23% of cell dry weight, a substrate to cell conversion rate of 0.13 g/g, and an oil yield of 0.03 g/g substrate [
115].
Aromatic compounds are the preferred destination for high-value bioremediation of benzene based plastic degradation products. Hee et al. [
98] obtained the engineered strain HBH-1 by expressing TphAabc and TphB from
Comamonas sp. E6 in and out of
Escherichia coli, and first achieved the conversion of phthalic acid to protocatechuic acid. Furthermore, Hee et al. conducted a series of high-value aromatic chemical synthesis studies using protocatechuic acid as a precursor. Firstly, by exogenous expression of p-hydroxybenzoate hydroxylase (PobA) from
P. putida KT2440,
Escherichia coli GA-1 was able to convert protocatechuic acid into 1.4 mmol/L gallic acid with a conversion rate of 40.1%. To eliminate the imbalance of cofactors in the synthesis of gallic acid by a single bacterium, Hee et al. divided the synthesis pathway of gallic acid into two modules: the protocatechuic acid synthesis module (PCA-1) and the gallic acid synthesis module (HBH-2). Under the optimal strain inoculation conditions, the conversion rate of the system from protocatechuic acid to gallic acid reached 92.5%. Using the same strategy, by exogenous expression of gallic acid decarboxylase (Lpdc) in the gallic acid synthesis strain GA-1, the engineered strain PG-1a can achieve 32.7% conversion of TPA to pyrogallol. To address the accumulation of by-products such as catechins during this conversion process, Hee et al. constructed another pathway for the synthesis of catechins using catechins as an intermediate: the original catechins undergo decarboxylation to form catechol, which is then converted into catechol through the action of phenol hydroxylase (PhKLMNOPQ). By co-culturing strains for catechin synthesis and catechol synthesis, the final yield of catechol synthesis from TPA reached 0.6 mmol/L, which is three times that of single strain cultivation. Using the same strategy, Hee et al. also completed the synthesis of TPA to viscous citric acid and vanillic acid, providing valuable experience for the high-value recovery of PET degradation products.
Many plastic degradation products or their intermediate metabolites have cytotoxicity, severely inhibiting cell growth and product synthesis. For example, the intermediate metabolites ethanal and glyoxal are important intermediates in the metabolism of EG, and 4 mmol/L ethanal and 7.5 mol/L EG can completely inhibit the growth of
Pseudomonas aeruginosa [
90]. Accelerating the conversion of aldehydes to corresponding less toxic alcohols or acids is a common strategy for reducing the toxicity of aldehydes. Based on this, Franden et al. [
91] reduced the accumulation of ethanal by overexpressing the glycolate oxidase GlcDEF, allowing the
P. putida engineering bacterium MFL114 to tolerate 2 mol/L (approximately 124 g/L) of EG and ultimately consume 0.5 mol/L (31 g/L) of EG to generate PHA with a dry cell weight of 32.19%.
The degradation products of plastics are diverse in composition, and a single microorganism is often insufficient to completely degrade them. The use of mixed microbial cultures, with targeted selection of functional microorganisms, may accelerate the biorefining process of plastic degradation products. The main degradation monomers of PU (polyurethane) plastics include adipic acid, EG, 1,4-butanediol and isocyanates (such as toluene-2,4-diisocyanate (2,4-TDI) or 4,4′-methylenediphenyl diisocyanate (4,4′-MDI)). Isocyanates further derive and transform into 2,4-toluenediamine. Based on this, researchers first identified and constructed microorganisms capable of degrading polyurethane monomers [
116]. It has been reported that
Acinetobacter beijingii ADP1 has excellent adipic acid degradation ability. By cloning the key adipic acid degradation gene cluster (dac: dcaAKIJP) and expressing it exogenously in
P. putida KT2440, the engineered strain
P. putida KT2440 A12.1p acquired the ability to grow rapidly in a medium where adipic acid is the carbon source. Mutant strains
P. putida KT2440 B10.1 and engineered strain
P. putida KT2440 ΔgclRΔPP_2046 ΔPP_2662::14d could degrade 1,4-butanediol and EG, respectively. The study also found that 2,4-toluenediamine exhibited significant cytotoxicity, which negatively impacted the bioavailability of other monomers in the polyurethane hydrolysis system. Therefore, researchers used paraffin oil and di(2-ethylhexyl) phosphoric acid (D2EHPA) as solvents and reactive extractants (co-solvents) to extract and remove toluenediamine from the polyurethane hydrolysis system. Under pH 4 conditions, the TDA removal efficiency reached 93%. Under these conditions, through mixed cultivation of
P. putida KT2440 A12.1p and
P. putida KT2440 B10.1.
5. Conclusions and Prospects
In recent years, researchers from domestic and international have carried out various screening of microbial resources for plastic waste degradation and excavation and modification of key enzyme components, achieving remarkable breakthroughs in the enzymatic depolymerization and catalytic mechanism of polyester plastics such as PET [
76,
117,
118]. The enzymatic recycling technology of CARBIOS has been industrialized in France, and it is expected to build the world’s first PET biological recycling plant by the end of 2025, with an annual processing capacity of 50,000 tons of post consumer PET waste. These achievements fully demonstrate that bio-depolymerization technology based on synthetic biology will be an effective way to achieve the recycling of waste plastic resources. However, there are many types and complex components of waste plastics, and the biodegradation of other waste plastics besides PET plastics still faces a series of bottleneck problems such as a lack of degradation bacteria/enzymes, unclear depolymerization mechanisms, low degradation system efficiency, and difficult efficient utilization of degradation products, which urgently need to be overcome.
5.1. Biological Oxidation Mechanism of Non Hydrolytic Plastics
Non-hydrolyzable plastics (such as PE, PP, PVC, and PS) account for more than 60% of the market share. These plastics have chemical structures with abundant inert C-C backbones and lack other reactive functional groups, which makes them resistant to enzymatic attack and can only degrade through high-energy oxidation reactions [
119]. Recently, some successful reports have been made on the depolymerization of non-hydrolyzable plastics by insects and their gut microbiota, but the biological degradation efficiency remains relatively low, and the biological oxidase and catalytic mechanisms involved in their degradation process still need further clarification [
62,
63,
120,
121]. Since polyethylene and straight-chain alkanes share the same monomer structure, it is speculated that their biodegradation mechanisms are similar. Alkane hydroxylases/monooxygenases (AlkB) are considered as candidate enzymes involved in polyethylene degradation. Lignin-degrading enzymes (such as LMSs) are effective in degrading aromatic structures and may be potential degraders of the aromatic plastic polystyrene. Therefore, based on the molecular catalytic mechanisms of alkane hydroxylases/monooxygenases and lignin-degrading enzymes, a “bottom-up” design approach for biologically oxidizing enzymes guided by structure might be more suitable than the “top-down” insect degradation approach to uncover the biological oxidation mechanisms of non-hydrolyzable plastics.
5.2. Efficient Heterologous Expression of Plastic Depolymerase
The efficient and low-cost preparation of plastic depolymerase formulations is crucial for the industrial-scale demonstration of biological plastic degradation. Currently, the soluble expression levels of plastic depolymerases are generally low in host organisms such as
Escherichia coli,
Pichia pastoris, and
Bacillus subtilis. For example, the soluble expression level of bacterial PETase is 300 mg/L, while the soluble expression level of fungal cutinase TfCa is 6.3 mg/L. The soluble expression level of the metagenomic polyester hydrolase LCC is approximately 40 mg/L. The expression levels of redox enzymes such as laccase, peroxidase, and P450 enzymes are even lower. During the heterologous expression of plastic depolymerases, when the solute-solute (protein-protein) interactions are stronger than the solute-solvent (protein-buffer) interactions, the increased protein-protein interactions (such as electrostatic interactions and hydrophobic forces) lead to protein aggregation, which is a key factor affecting their efficient expression. With the advancement of synthetic biology, protein engineering technologies aimed at improving the colloidal and structural stability of proteins, as well as post-translational modification strategies to enhance protein glycosylation, are expected to provide an effective solution to the protein aggregation problem in plastic depolymerases from the source.
5.3. Construction of a Multi Enzyme/Mixed Bacterial System for Depolymerization of Mixed Plastics
For plastics with complex structures (such as PU) or mixed plastics, a single degrading microorganism/enzyme is often difficult to achieve effective degradation. To reduce the metabolic pressure of a single microorganism, the degradation process can be divided into multiple different processes to complete. The bacterium
Ideonella sakaiensis 201-F6 achieves sufficient degradation of PET through the synergistic secretion of highly active PETase and MHETase [
76]; Chen et al. [
122] also achieved in-situ biodegradation of mixed microplastics in activated sludge based on extreme thermophilic composting technology. The multi-enzyme/mixed bacterial degradation system has achieved good results in the field of waste carbon resource degradation, such as the biodegradation of lignocellulose [
123]. Compared with lignocellulose, plastics have higher hydrophobicity and crystallinity, and stronger resistance to biodegradation [
124]. Therefore, based on the acquisition of various plastic degrading microorganisms and degradation elements, designing and constructing efficient and stable multi-enzyme/mixed bacterial systems, and directing the dynamic changes of multi-enzyme/mixed bacterial systems according to different types of plastic compositions will be the key breakthrough direction for plastic biodegradation.
5.4. High-Value Biorefinery Pathway for Plastic Degradation Products
Establishing a biological utilization pathway of “plastic waste depolymerization monomers high-value products” can not only promote the development of circular economy in the plastic industry, but also effectively save natural resources, reduce greenhouse gas emissions, and protect the ecological environment. The European Union launched the EU Horizon 2020 Plastic Biodegradation and Utilization Project P4SB (from plastic waste to plastic value using Pseudomonas putida synthetic biology) as early as 2015, which connected the technical route of synthesizing biodegradable plastic PHA and biosurfactant rhamnose ester from PET plastic monomers EG/TPA and PUR plastic monomers butanediol/adipic acid [
91,
94,
125,
126], forming a top international research team in the field of plastic degradation (https://www.p4sb.eu/ (accessed on 15 January 2025)). However, the efficiency of existing high-value biorefinery pathways is still relatively low [
127]. Designing and constructing efficient plastic monomer degradation and assimilation pathways, and regulating the compatibility between the two pathways and chassis cells, truly achieving “plastic reduction and recycling” and promoting the development of circular economy, synthetic biotechnology will play an important role.
Given the serious issues of “white pollution” and the waste of plastic resources, the Chinese 13th Five-Year Plan for Science and Technology has placed a high priority on research into plastic biodegradation and conversion. In 2019, the National Natural Science Foundation of China (NSFC) and the European Commission jointly funded two high-intensity international (regional) cooperation and exchange projects in the field of “plastic degradation microorganisms. The “Microbial Communities for Synthetic Plastic Degradation and Conversion” project was led by Professor Qingsheng Qi from Shandong University and Professor Margaret Brennan Fournet from the Athlone Institute of Technology in Ireland (https://www.bioicep.eu/ (accessed on 8 January 2025)). The “Key Scientific Issues and Technologies for Efficient Biodegradation and Conversion of Waste Plastics” project was led by Professor Min Jiang from Nanjing University of Technology and Professor Lars M. Blank from Aachen University of Technology (https://www.mix-up.eu/ (accessed on 16 January 2025)). In the same year, the Ministry of Science and Technology (MOST) launched two key research areas in the National Key R&D Program: “Key Scientific Issues of Transformational Technologies” and “Synthetic Biology.” These areas included the “Directed Evolution Engineering and Application of Synthetic Plastic Degrading Enzymes” and “Artificial Multicellular Systems Construction and Application in Activated Sludge” projects, which were funded for Professor Wu Jing from Jiangnan University and Researcher Liu Shuangjiang from the Institute of Microbiology, Chinese Academy of Sciences. In 2024, the progress meeting for the National Key R&D Program “Green Bio-Manufacturing” project on “Development and Industrialization Demonstration of Bio-based Polyurethane Polyol New Products and Their Green Manufacturing Technologies” was held in Nanjing. With the advancement of synthetic biology and its widespread application in plastic biodegradation and conversion, it is believed that China will make exciting research progress in the biological degradation and conversion of waste plastic resources.
Author Contributions
Conceptualization, W.D. and X.Q.; Validation, X.Q., M.W. and F.Z.; Writing—Original Draft Preparation, M.W. and X.H.; Writing—Review & Editing, M.W. and J.Z.; Supervision, X.Q.; Project Administration, M.J., Q.L.
Ethics Statement
Not applicable.
Informed Consent Statement
Not applicable.
Funding
This work was supported by the National Key R & D Program of China (2024YFC3908300), the National Natural Science Foundation of China (22478184, 22408168), Jiangsu Basic Research Center for Synthetic Biology (BK20233003), and the Open Project Program of Fujian Huafeng New Material Co., Ltd. (SSUM243).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
1.
Adyel TM. Accumulation of plastic waste during COVID-19.
Science 2020,
369, 1314–1315.
[Google Scholar]
2.
Samantaray PK, Little A, Haddleton DM, McNally T, Tan B, Sun Z, et al. Poly(glycolic acid) (PGA), a versatile building block expanding high performance and sustainable Bioplastic applications.
Green Chem. 2020,
22, 4055–4081.
[Google Scholar]
3.
Geyer R, Jambeck JR, Law KL. Production, use, and fate of all plastics ever made.
Sci. Adv. 2017,
3, e1700782.
[Google Scholar]
4.
Stubbins A, Law KL, Muñoz SE, Bianchi TS, Zhu L. Plastics in the Earth system.
Science 2021,
373, 51–55.
[Google Scholar]
5.
Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, et al. Marine pollution. Plastic waste inputs from land into the ocean.
Science 2015,
347, 768–771.
[Google Scholar]
6.
Stevens DR, Bommarito PA, Keil AP, McElrath TF, Trasande L, Barrett ES, et al. Urinary phthalate metabolite mixtures in pregnancy and fetal growth: Findings from the infant development and the environment study.
Environ. Int. 2022,
163, 107235.
[Google Scholar]
7.
Sen GR, Samantaray PK, Bose S. Going beyond Cellulose and Chitosan: Synthetic Biodegradable Membranes for Drinking Water, Wastewater, and Oil-Water Remediation.
ACS Omega 2023,
8, 24695–24717.
[Google Scholar]
8.
Undas AK, Groenen M, Peters RJB, van Leeuwen SPJ. Safety of recycled plastics and textiles: Review on the detection, identification and safety assessment of contaminants.
Chemosphere 2023,
312, 137175.
[Google Scholar]
9.
Thew C, Lee ZS, Srinophakun P, Ooi CW. Recent advances and challenges in sustainable management of plastic waste using biodegradation approach.
Bioresour. Technol. 2023,
374, 128772.
[Google Scholar]
10.
Schneier A, Melaugh G, Sadler JC. Engineered plastic-associated bacteria for biodegradation and bioremediation.
Biotechnol. Environ. 2024,
1, 7.
[Google Scholar]
11.
Thomsen TB, Almdal K, Meyer AS. Significance of poly(ethylene terephthalate) (PET) substrate crystallinity on enzymatic degradation.
New Biotechnol. 2023,
78, 162–172.
[Google Scholar]
12.
Aarsen CV, Liguori A, Mattsson R, Sipponen MH, Hakkarainen M. Designed to Degrade: Tailoring Polyesters for Circularity.
Chem. Rev. 2024,
124, 8473–8515.
[Google Scholar]
13.
Araújo R, Silva C, O'Neill A, Micaelo N, Guebitz G, Soares CM, et al. Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers.
J. Biotechnol. 2007,
128, 849–857.
[Google Scholar]
14.
Nimchua T, Eveleigh DE, Sangwatanaroj U, Punnapayak H. Screening of tropical fungi producing polyethylene terephthalate-hydrolyzing enzyme for fabric modification.
J. Ind. Microbiol. Biotechnol. 2008,
35, 843–850.
[Google Scholar]
15.
Ronkvist SA, Xie W, Lu WH, Richard A. Cutinase catalyzed hydrolysis of poly(ethylene terephthalate).
Macromolecules 2009,
42, 5128–5138.
[Google Scholar]
16.
Kleeberg I, Hetz C, Kroppenstedt RM, Müller RJ, Deckwer WD. Biodegradation of aliphatic-aromatic copolyesters by Thermomonospora fusca and other thermophilic compost isolates.
Appl. Environ. Microbiol. 1998,
64, 1731–1735.
[Google Scholar]
17.
Muller RJ, Schrader H, Profe J, Dresler K, Deckwer WD. Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca.
Macromol. Rapid Commun. 2010,
26, 1400–1405.
[Google Scholar]
18.
Then J, Wei R, Oeser T, Gerdts A, Schmidt J, Barth M, et al. A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate.
FEBS Open Bio. 2016,
6, 425–432.
[Google Scholar]
19.
Then J, Wei R, Oeser T, Barth M, Belisário-Ferrari MR, Schmidt J, et al. Ca
2+ and Mg
2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca.
Biotechnol. J. 2015,
10, 592–598.
[Google Scholar]
20.
Hu XP, Osaki S, Hayashi M, Kaku M, Katuen S, Kobayashi H, et al. Degradation of a terephthalate-containing polyester by Thermophilic Actinomycetes and Bacillus species derived from composts.
J. Polym. Environ. 2008,
16, 103–108.
[Google Scholar]
21.
Srivastava P, Saji J, Manickam N. Biodegradation of polyethylene terephthalate (PET) by Brucella intermedia IITR130 and its proposed metabolic pathway.
Biodegradation 2024,
35, 671–685.
[Google Scholar]
22.
Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, et al. A bacterium that degrades and assimilates poly(ethylene terephthalate).
Science 2016,
351, 1196–1199.
[Google Scholar]
23.
Chandramouli Swamy TM, Nagarathna SV, Reddy PV, Nayak AS. Efficient biodegradation of Polyethylene terephthalate (PET) plastic by Gordonia sp. CN2K isolated from plastic contaminated environment.
Ecotoxicol. Environ. Saf. 2024,
281, 116635.
[Google Scholar]
24.
Sui B, Wang T, Fang J, Hou Z, Shu T, Lu Z, et al. Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes.
Front. Microbiol. 2023,
14, 1265139.
[Google Scholar]
25.
Kleeberg I, Welzel K, Vandenheuvel J, Müller RJ, Deckwer WD. Characterization of a new extracellular hydrolase from Thermobifida fusca degrading aliphatic-aromatic copolyesters.
Biomacromolecules 2005,
6, 262–270.
[Google Scholar]
26.
Feder D. Humicola Insolens Cutinase; A Novel Catalyst for Polymer Synthesis Reactions. Ph.D. Thesis, Polytechnic Institute of New York University: New York, NY, USA, 2013.
27.
Sulaiman S, You DJ, Kanaya E, Koga Y, Kanaya S. Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
Biochemistry 2014,
25, 1858–1869.
[Google Scholar]
28.
Makryniotis K, Nikolaivits E, Gkountela C, Vouyiouka S, Topakas E. Discovery of a polyesterase from Deinococcus maricopensis and comparison to the benchmark LCCICCG suggests high potential for semicrystalline post-consumer PET degradation.
J. Hazard. Mater. 2023,
455, 131574.
[Google Scholar]
29.
Mahajan N, Gupta P. New insights into the microbial degradation of polyurethanes.
RSC Adv. 2015,
5, 41839–41854.
[Google Scholar]
30.
Álvarez-Barragán J, Domínguez-Malfavón L, Vargas-Suárez M, González-Hernández R, Aguilar-Osorio G, Loza-Tavera H. Biodegradative Activities of Selected Environmental Fungi on a Polyester Polyurethane Varnish and Polyether Polyurethane Foams.
Appl. Environ. Microbiol. 2016,
82, 5225–5235.
[Google Scholar]
31.
Mathur G, Prasad R. Degradation of polyurethane by Aspergillus flavus (ITCC 6051) isolated from soil.
Appl. Biochem. Biotechnol. 2012,
167, 1595–1602.
[Google Scholar]
32.
Osman M, Satti SM, Luqman A, Hasan F, Shah Z, Shah AA. Degradation of polyester polyurethane by Aspergillus sp. strain S45 isolated from soil.
J. Polym. Environ. 2018,
26, 301–310.
[Google Scholar]
33.
Khan S, Nadir S, Shah ZU, Shah AA, Karunarathna SC, Xu J, et al. Biodegradation of polyester polyurethane by Aspergillus tubingensis.
Environ. Pollut. 2017,
225, 469–480.
[Google Scholar]
34.
Rajan A, Ameen F, Jambulingam R, Shankar V. Biodegradation of Polyurethane by Fungi Isolated from Industrial Wastewater
—A Sustainable Approach to Plastic Waste Management.
Polymers 2024,
16, 1411.
[Google Scholar]
35.
Pantelic B, Siaperas R, Budin C, de Boer T, Topakas E, Nikodinovic-Runic J. Proteomic examination of polyester-polyurethane degradation by Streptomyces sp. PU10: Diverting polyurethane intermediates to secondary metabolite production.
Microb. Biotechnol. 2024,
17, e14445.
[Google Scholar]
36.
Magnin A, Pollet E, Phalip V, Avérous L. Evaluation of biological degradation of polyurethanes.
Biotechnol. Adv. 2020,
39, 107457.
[Google Scholar]
37.
Ji J, Pei J, Ding F, Zeng C, Zhou J, Dong W, et al. Isolation and characterization of polyester polyurethane-degrading bacterium
Bacillus sp. YXP1.
Environ. Res. 2024,
15, 118468.
[Google Scholar]
38.
Peng R, Xia M, Ru J, Huo Y, Yang Y. Microbial degradation of polyurethane plastics.
Chin. J. Biotechnol. 2018,
34, 1398–1409.
[Google Scholar]
39.
Akutsu Y, Nakajima-Kambe T, Nomura N, Nakahara T. Purification and Properties of a Polyester Polyurethane-Degrading Enzyme from Comamonas acidovorans TB-35.
Appl. Environ. Microbiol. 1998,
64, 2–7.
[Google Scholar]
40.
Nomura N, Shigeno-Akutsu Y, Nakajima-Kambe T, Nakahara T. Cloning and sequence analysis of a polyurethane esterase of Comamonas acidovorans TB-35.
J. Ferment. Bioeng. 1998,
86, 339–345.
[Google Scholar]
41.
Stern RV, Howard GT. The polyester polyurethanase gene (pueA) from Pseudomonas chlororaphis encodes a lipase.
FEMS Microbiol. Lett. 2000,
185, 163–168.
[Google Scholar]
42.
Howard GT, Crother B, Vicknair J. Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas chlororaphis.
Int. Biodeter. Biodegr. 2001,
47, 141–149.
[Google Scholar]
43.
Ru J, Chen X, Dong X, Hu L, Zhang J, Yang Y. Discovery of a polyurethane-degrading enzyme from the gut bacterium of plastic-eating mealworms.
J. Hazard. Mater. 2024,
480, 136159.
[Google Scholar]
44.
Phua SK, Castillo E, Anderson JM, Hiltner A. Biodegradation of a polyurethane
in vitro.
J. Biomed. Mater. Res. 1987,
21, 231–246.
[Google Scholar]
45.
Campiñez MD, Aguilar-de-Leyva Á, Ferris C, de Paz MV, Galbis JA, Caraballo I. Study of the properties of the new biodegradable polyurethane PU (TEG-HMDI) as matrix forming excipient for controlled drug delivery.
Drug Dev. Ind. Pharm. 2013,
39, 1758–1764.
[Google Scholar]
46.
Branson Y, Söltl S, Buchmann C, Wei R, Schaffert L, Badenhorst CPS, et al. Urethanases for the enzymatic hydrolysis of low molecular weight carbamates and the recycling of polyurethanes.
Angew. Chem. Int. Ed. 2023,
62, e202216220.
[Google Scholar]
47.
Schmidt J, Wei R, Oeser T, Dedavid E, Silva LA, Breite D, et al. Degradation of polyester polyurethane by bacterial polyester hydrolases.
Polymers 2017,
9, 65.
[Google Scholar]
48.
Roman VA, Crable BR, Wagner DN, Gryganskyi A, Zelik S, Cummings L, et al. Identification and recombinant expression of a cutinase from Papiliotrema laurentii that hydrolyzes natural and synthetic polyesters.
Appl. Environ. Microbiol. 2024,
90, e0169423.
[Google Scholar]
49.
Liu J, Xin K, Zhang T, Wen Y, Li D, Wei R, et al. Identification and characterization of a fungal cutinase-like enzyme CpCut1 from Cladosporium sp. P7 for polyurethane degradation.
Appl. Environ. Microbiol. 2024,
90, e0147723.
[Google Scholar]
50.
Ruiz C, Howard GT. Nucleotide sequencing of a polyurethanase gene (pulA) from Pseudomonas fluorescens.
Int. Biodeter. Biodegr. 1999,
44, 127–131.
[Google Scholar]
51.
Xu Y, Yin CF, Yue WL, Zhou NY. Microbial degradation of petroleum-based plastics.
Chin. J. Biotechnol. 2019,
35, 2092–2103.
[Google Scholar]
52.
Han QX, Wang QZ, Zhang M. Study on biodegradability of modified PE film.
Chin. J. Plast. Ind. 2009,
37, 48–51.
[Google Scholar]
53.
Balasubramanian V, Natarajan K, Hemambika B, Ramesh N, Sumathi CS, Kottaimuthu R, et al. High-density polyethylene (HDPE)-degrading potential bacteria from marine ecosystem of Gulf of Mannar, India.
Lett. Appl. Microbiol. 2010,
51, 205–211.
[Google Scholar]
54.
Tribedi P, Sil AK. Low-density polyethylene degradation by
Pseudomonas sp. AKS2 biofilm.
Environ. Sci. Pollut. Res. Int. 2013,
20, 4146–4153.
[Google Scholar]
55.
Gilan I, Hadar Y, Sivan A. Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber.
Appl. Microbiol. Biotechnol. 2004,
65, 97–104.
[Google Scholar]
56.
Awasthi S, Srivastava P, Singh P, Tiwary D, Mishra P. Biodegradation of thermally treated high-density polyethylene (HDPE) by Klebsiella pneumoniae CH001.
3 Biotech 2017,
7, 332.
[Google Scholar]
57.
Kavitha R, Bhuvaneswari V. Assessment of polyethylene degradation by biosurfactant producing ligninolytic bacterium.
Biodegradation 2021,
32, 531–549.
[Google Scholar]
58.
Pathak VM, Navneet. Exploitation of bacterial strains for microplastics (LDPE) biodegradation.
Chemosphere 2023,
316, 137845.
[Google Scholar]
59.
Sun W, Zhang Y, Zhang H, Wu H, Liu Q, Yang F, et al. Exploitation of Enterobacter hormaechei for biodegradation of multiple plastics.
Sci. Total Environ. 2024,
907, 167708.
[Google Scholar]
60.
Eisaku KT, Linn E, Takeshi O, Taneaki I, Ishibashi Y. Isolation and characterization of polystyrene degrading microorganisms for zero emission treatment of expanded polystyrene.
Environ. Eng. Res. 2003,
40, 373–379.
[Google Scholar]
61.
Tian L, Kolvenbach B, Corvini N, Wang S, Tavanaie N, Wang L, et al. Mineralisation of 14C-labelled polystyrene plastics by Penicillium variabile after ozonation pre-treatment.
New Biotechnol. 2017,
38, 101–105.
[Google Scholar]
62.
Yang Y, Yang J, Wu WM, Zhao J, Song Y, Gao L, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms (I): Chemical and physical characterization and isotopic tests.
Environ. Sci. Technol. 2015,
49, 12080–12086.
[Google Scholar]
63.
Yang Y, Yang J, Wu WM, Zhao J, Song Y, Gao L, et al. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms.
Environ. Sci. Technol. 2014,
48, 13776–13784.
[Google Scholar]
64.
Yang Y, Yang J, Wu WM, Zhao J, Song Y, Gao L, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms (Ⅱ): Role of gut microorganisms.
Environ. Sci. Technol. 2015,
49, 12087–12093.
[Google Scholar]
65.
Liu R, Zhao S, Zhang B, Li G, Fu X, Yan P, et al. Biodegradation of polystyrene (PS) by marine bacteria in mangrove ecosystem.
J. Hazard. Mater. 2023,
442, 130056.
[Google Scholar]
66.
Park JW, Kim M, Kim SY, Bae J, Kim TJ. Biodegradation of polystyrene by intestinal symbiotic bacteria isolated from mealworms, the larvae of Tenebrio molitor.
Heliyon 2023,
9, e17352.
[Google Scholar]
67.
Miravalle E, Balboa S, Zanetti M, Otero A, Lazzari M. New insights on the degradation of polystyrene and polypropylene by larvae of the superworm Zophobas atratus and gut bacterial consortium enrichments obtained under different culture conditions.
J. Hazard. Mater. 2024,
478, 135475.
[Google Scholar]
68.
Wei R, Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we?
Microb. Biotechnol. 2017,
10, 1308–1322.
[Google Scholar]
69.
Santo M, Weitsman R, Sivan A. The role of the copper binding enzyme-laccase-in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber.
Int. Biodeter. Biodegrad. 2013,
84, 204–210.
[Google Scholar]
70.
Fujisawa M, Hirai H, Nishida T. Degradation of polyethylene and Nylon-66 by the laccase-mediator system.
J. Polym. Environ. 2001,
9, 103–108.
[Google Scholar]
71.
Rojo F. Degradation of alkanes by bacteria.
Environ. Microbiol. 2009,
11, 2477–2490.
[Google Scholar]
72.
Yoon M, Jeon H, Kim M. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell.
J. Bioremed. Biodegrad. 2012,
3, 8.
[Google Scholar]
73.
Jeon H, Kim M. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation.
Int. Biodeter. Biodegrad. 2015,
103, 141–146.
[Google Scholar]
74.
Nakamiya K, Sakasita G, Ooi T, Kinoshita S. Enzymatic degradation of polystyrene by hydroquinone peroxidase of Azotobacter beijerinckii HM121.
J. Ferment. Bioeng. 1997,
84, 480–482.
[Google Scholar]
75.
Arnling Bååth J, Novy V, Carneiro LV, Guebitz GM, Olsson L, Westh P, et al. Structure-function analysis of two closely related cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng. 2022,
119, 470–481.
[Google Scholar]
76.
Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, et al. An engineered PET depolymerase to break down and recycle plastic bottles.
Nature 2020,
580, 216–219.
[Google Scholar]
77.
Wei R, Oeser T, Barth M, Weigl N, Lübs A, Schulz-Siegmund M, et al. Turbidimetric analysis of the enzymatic hydrolysis of polyethylene terephthalate nanoparticles.
J. Mol. Catal. B Enzym. 2014,
103, 72–78.
[Google Scholar]
78.
Ribitsch D, Herrero Acero E, Przylucka A, Zitzenbacher S, Marold A, Gamerith C, et al. Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins.
Appl. Environ. Microbiol. 2015,
81, 3586–3592.
[Google Scholar]
79.
Ribitsch D, Yebra AO, Zitzenbacher S, Wu J, Nowitsch S, Steinkellner G, et al. Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis.
Biomacromolecules 2013,
14, 1769–1776.
[Google Scholar]
80.
Gamerith C, Acero EH, Pellis A, Ortner A, Vielnascher R, Luschnig D, et al. Improving enzymatic polyurethane hydrolysis by tuning enzyme sorption.
Polym. Degrad. Stabil. 2016,
132, 69–77.
[Google Scholar]
81.
Silva C, Da S, Silva N, Matamá T, Araújo R, Martins M, et al. Engineered Thermobifida fusca cutinase with increased activity on polyester substrates.
Biotechnol. J. 2011,
6, 1230–1239.
[Google Scholar]
82.
Zheng Y, Li QB, Liu P, Yuan YB, Dian LY, Wang Q, et al. Dynamic Docking-Assisted engineering of hydrolases for efficient PET depolymerization.
ACS Catal. 2024,
5, 3627–3639.
[Google Scholar]
83.
Cui Y, Chen Y, Sun J, Zhu T, Pang H, Li C, et al. Computational redesign of a hydrolase for nearly complete PET depolymerization at industrially relevant high-solids loading.
Nat. Commun. 2024,
15, 1417.
[Google Scholar]
84.
Li Z, Han X, Cong L, Singh P, Paiva P, Branson Y, et al. Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv. Sci. 2025,
7, e2416019.
[Google Scholar]
85.
Wei R, Oeser T, Schmidt J, Meier R, Barth M, Then J, et al. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition.
Biotechnol. Bioeng. 2016,
113, 1658–1665.
[Google Scholar]
86.
Carniel A, Valoni E, Nicomedes Junior J, Gomes ADC, Castro AMd. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid.
Process Biochem. 2017,
59, 84–90.
[Google Scholar]
87.
Barth M, Honak A, Oeser T, Wei R, Belisário-Ferrari MR, Then J, et al. A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films.
Biotechnol. J. 2016,
11, 1082–1087.
[Google Scholar]
88.
Zhang J, Wang H, Luo Z, Yang Z, Zhang Z, Wang P, et al. Computational design of highly efficient thermostable MHET hydrolases and dual enzyme system for PET recycling.
Commun. Biol. 2023,
6, 1135.
[Google Scholar]
89.
Parke D, Garcia MA, Ornstin LN. Cloning and genetic characterization of dca genes required for β-oxidation of straight-chain dicarboxylic acids in Acinetobacter sp. strain ADP1.
Appl. Environ. Microbiol. 2001,
67, 4817–4827.
[Google Scholar]
90.
Choi JH, Kim TK, Kim YM, Kim WC, Park K, Rhee I. Cloning and characterization of a gene cluster for cyclohexanone oxidation in Rhodococcus sp. TK6.
J. Microbiol. Biotechnol. 2006,
16, 511–518.
[Google Scholar]
91.
Franden MA, Jayakody LN, Li WJ, Wagner NJ, Cleveland NS, Michener WE, et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization.
Metab. Eng. 2018,
48, 197–207.
[Google Scholar]
92.
Mückschel B, Simon O, Klebensberger J, Graf N, Rosche B, Altenbuchner J, et al. Ethylene glycol metabolism by Pseudomonas putida.
Appl. Environ. Microbiol. 2012,
78, 8531–8539.
[Google Scholar]
93.
Trifunović D, Schuchmann K, Müller V. Ethylene Glycol Metabolism in the Acetogen Acetobacterium woodii.
J. Bacteriol. 2016,
198, 1058–1065.
[Google Scholar]
94.
Li WJ, Narancic T, Kenny ST, Niehoff PJ, O'Connor K, Blank LM, et al. Unraveling 1,4-Butanediol Metabolism in Pseudomonas putida KT2440.
Front. Microbiol. 2020,
11, 382.
[Google Scholar]
95.
Sasoh M, Masai E, Ishibashi S, Hara H, Kamimura N, Miyauchi K, et al. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6.
Appl. Environ. Microbiol. 2006,
72, 1825–1832.
[Google Scholar]
96.
Shigematsu T, Yumihara K, Ueda Y, Morimura S, Kida K. Purification and gene cloning of the oxygenase component of the terephthalate 1,2-dioxygenase system from Delftia tsuruhatensis strain T7.
FEMS Microbiol. Lett. 2003,
220, 255–260.
[Google Scholar]
97.
Choi KY, Kim D, Sul WJ, Chae JC, Zylstra GJ, Kim YM, et al. Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcus sp. strain DK17.
FEMS Microbiol. Lett. 2005,
252, 207–213.
[Google Scholar]
98.
Kim HT, Kim JK, Cha HG, Kang MJ, Lee SH, Khang MJ, et al. Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET.
ACS Sustain. Chem. Eng. 2019,
7, 19396–19406.
[Google Scholar]
99.
Maurya AC, Bhattacharya A, Khare SK. Biodegradation of terephthalic acid using Rhodococcus erythropolis MTCC 3951: Insights into the degradation process, applications in wastewater treatment and polyhydroxyalkanoate production.
Environ. Sci. Pollut. Res. Int. 2024,
31, 57376–57385.
[Google Scholar]
100.
Kenny ST, Runic JN, Kaminsky W, Woods T, Babu RP, O'Connor KE. Development of a bioprocess to convert PET derived terephthalic acid and biodiesel derived glycerol to medium chain length polyhydroxyalkanoate.
Appl. Microbiol. Biotechnol. 2012,
95, 623–633.
[Google Scholar]
101.
Espinosa MJC, Blanco AC, Schmidgall T, Atanasoff-Kardjalieff AK, Kappelmeyer U, Tischler D, et al. Toward Biorecycling: Isolation of a Soil Bacterium That Grows on a Polyurethane Oligomer and Monomer.
Front. Microbiol. 2020,
11, 404.
[Google Scholar]
102.
Tenagy, Park JS, Iwama R, Kobayashi S, Ohta A, Horiuchi H, et al. Involvement of acyl-CoA synthetase genes in n-alkane assimilation and fatty acid utilization in yeast Yarrowia lipolytica.
FEMS Yeast Res. 2015,
15, fov031.
[Google Scholar]
103.
Patrauchan MA, Florizone C, Eapen S, Gómez-Gil L, Sethuraman B, Fukuda M, et al. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1.
J. Bacteriol. 2008,
190, 37–47.
[Google Scholar]
104.
Bestetti G, Di Gennaro P, Colmegna A, Ronco I, Galli E, Sello G. Characterization of styrene catabolic genes of Pseudomonas putida SN1 and construction of a recombinant Escherichia coli containing styrene monooxygenase gene for the production of (S)-styreneoxide.
J. Microbiol. Biotechnol. 2006,
16, 1032–1040.
[Google Scholar]
105.
Itoh N, Yoshida K, Okada K. Isolation and identification of styrene-degrading Corynebacterium strains, and their styrene metabolism.
Biosci. Biotechnol. Biochem. 1996,
60, 1826–1830.
[Google Scholar]
106.
Toda H, Itoh N. Isolation and characterization of styrene metabolism genes from styrene-assimilating soil bacteria Rhodococcus sp. ST-5 and ST-10.
J. Biosci. Bioeng. 2012,
113, 12–19.
[Google Scholar]
107.
Oelschlägel M, Zimmerling J, Tischler D. A review: the styrene metabolizing cascade of side-chain oxygenation as biotechnological basis to gain various valuable compounds.
Front. Microbiol. 2018,
9, 490.
[Google Scholar]
108.
Nikodinovic-Runic J, Casey E, Duane GF, Mitic D, Hume AR, Kenny ST, et al. Process analysis of the conversion of styrene to biomass and medium chain length polyhydroxyalkanoate in a two-phase bioreactor.
Biotechnol. Bioeng. 2011,
108, 2447–2455.
[Google Scholar]
109.
Sen SK, Raut S. Microbial degradation of low density polyethylene (LDPE): A review.
J. Environ. Chem. Eng. 2015,
3, 462–473.
[Google Scholar]
110.
Johnston B, Radecka I, Chiellini E, Barsi D, Ilieva VI, Sikorska W, et al. Mass spectrometry reveals molecular structure of polyhydroxyalkanoates attained by bioconversion of oxidized polypropylene waste fragments.
Polymers 2019,
11, 1580.
[Google Scholar]
111.
Ackermann YS, Li WJ, de Hipt LO, Niehoff PJ, Casey W, Polen T, et al. Engineering adipic acid metabolism in Pseudomonas putida.
Metab. Eng. 2021,
67, 29–40.
[Google Scholar]
112.
Kenny ST, Runic JN, Kaminsky W, Woods T, Babu RP, Keely CM, et al. Up-cycling of PET (polyethylene terephthalate) to the biodegradable plastic PHA (polyhydroxyalkanoate).
Environ. Sci. Technol 2008,
42, 7696–7701.
[Google Scholar]
113.
Christova N, Tuleva B, Lalchev Z, Jordanova A, Jordanov B. Rhamnolipid biosurfactants produced by Renibacterium salmoninarum 27BN during growth on n-hexadecane.
Z. Naturforsch. C J. Biosci. 2004,
59, 70–74.
[Google Scholar]
114.
Abdel-Mawgoud AM, Lepine F, Deziel E. A stereo-specific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants.
Chem. Biol. 2014,
21, 156–164.
[Google Scholar]
115.
Mihreteab M, Stubblefield BA, Gilbert ES. Microbial bioconversion of thermally depolymerized polypropylene by Yarrowia lipolytica for fatty acid production.
Appl. Microbiol. Biotechnol. 2019,
103, 7729–7740.
[Google Scholar]
116.
Catur Utomo RN, Li WJ, Tiso T, Eberlein C, Doeker M, Hepieper HJ, et al. Defined microbial mixed culture for utilization of polyurethane monomers.
ACS Sustain. Chem. Eng. 2020,
8, 17466–17474.
[Google Scholar]
117.
Lee GH, Kim DW, Jin YH, Kim SM, Lim ES, Cha MJ, et al. Biotechnological plastic degradation and valorization using systems metabolic engineering.
Int. J. Mol. Sci. 2023,
24, 15181.
[Google Scholar]
118.
Tumu K, Vorst K, Curtzwiler G. Global plastic waste recycling and extended producer responsibility laws.
J. Environ. Manag. 2023,
348, 119242.
[Google Scholar]
119.
Inderthal H, Tai SL, Harrison STL. Non-Hydrolyzable Plastics - An Interdisciplinary Look at Plastic Bio-Oxidation.
Trends Biotechnol. 2021,
39, 12–23.
[Google Scholar]
120.
Yang XG, Wen PP, Yang YF, Jia PP, Li WG, Pei DS. Plastic biodegradation by in vitro environmental microorganisms and
in vivo gut microorganisms of insects.
Front. Microbiol. 2023,
13, 1001750.
[Google Scholar]
121.
Barclay A, Acharya KR. Engineering Plastic Eating Enzymes Using Structural Biology.
Biomolecules 2023,
13, 1407.
[Google Scholar]
122.
Chen Z, Zhao W, Xing R, Xie S, Yang X, Cui P, et al. Enhanced in situ biodegradation of microplastics in sewage sludge using hyperthermophilic composting technology.
J. Hazard. Mater. 2020,
384, 121271.
[Google Scholar]
123.
Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, Lipzen A, et al. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes.
Science 2016,
351, 1192–1195.
[Google Scholar]
124.
Chen CC, Dai L, Ma L, Guo RT. Enzymatic degradation of plant biomass and synthetic polymers.
Nat. Rev. Chem. 2020,
4, 114–126.
[Google Scholar]
125.
Escapa IF, García JL, Bühler B, Blank LM, Prieto MA. The polyhydroxyalkanoate metabolism controls carbon and energy spillage in Pseudomonas putida.
Environ. Microbiol. 2012,
14, 1049–1063.
[Google Scholar]
126.
Wierckx N, Prieto MA, Pomposiello P, de Lorenzo V, O'Connor K, Blank LM. Plastic waste as a novel substrate for industrial biotechnology.
Microb. Biotechnol. 2015,
8, 900–903.
[Google Scholar]
127.
Ran J, Talebian-Kiakalaieh A, Zhang S, Hashem EM, Guo M, Qiao SZ. Recent advancement on photocatalytic plastic upcycling.
Chem. Sci. 2023,
15, 1611–1637.
[Google Scholar]