Contents
Efficient Extracellular Production of Phospholipase D in Escherichia coli via Genetic and Process Engineering Modification
Author Information
Other Information
Beijing Bioprocess Key Laboratory, State Key Laboratory of Chemical Resource Engineering, Beijing University of Chemical Technology, Beijing 100029, China
*
Authors to whom correspondence should be addressed.
Received: 08 February 2025 Accepted: 08 April 2025 Published: 14 April 2025
© 2025 The authors. This is an open access article under the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/).
Synth. Biol. Eng.
2025,
3(2), 10006;
DOI: 10.70322/sbe.2025.10006
ABSTRACT:
Phospholipase
D (PLD) is the key enzyme in the catalytic production of rare phospholipids
including phosphatidylserine. It was considered a promising method via genetic
manipulation for the heterologous production of PLD in the model chassis. Few works
focused on the extracellular production of PLD in engineered microbes. Herein,
genetic and process engineering modification strategies were developed to
achieve secretory production of PLD in Escherichia
coli. The N-terminal fusion secretion signal peptide OmpA and the plasmid
pBAD-gⅢC with pBAD promoter were proven to be the most effective in promoting
the secretory production of PLD. Given the limitation of the cell membrane, the
regulation of the key protein expression in the cell membrane as well as the
addition of surfactants, were explored to accelerate the secretory production
of PLD further. It was indicated that adding 0.5% (w/v) Triton
X-100 was more conducive to producing PLD. Finally, fed-batch fermentation was
conducted, and the maximum extracellular PLD activity achieved was 33.25 U/mL,
which was the highest level reported so far. Our work demonstrated the
effectiveness of genetic and process engineering strategies for the secretory
production of PLD in E. coli, which
provided an alternative platform for the industrial production of PLD.
Keywords:
Phospholipase D; Secretory
production; Signal peptide; Surfactant
1. Introduction
As a member of the phospholipase superfamily, phospholipase D (PLD) is a major membrane phospholipid-modifying enzyme in prokaryotes and eukaryotes. It can hydrolyze the phosphodiester bond of the substrate and catalyze the transphosphatidylation reaction to generate rare phospholipids, thus attracting much attention from researchers [1]. In particular, PLD from Streptomyces has a relatively high transphosphatidylation activity and can catalyze the transphosphatidylation reaction of L-serine to bind it to phosphatidylcholine to form phosphatidylserine (PS) [2].
As a functional food, PS can reduce oxidative stress in the brain and stimulate neurotransmitter release, which is known as a “brain-specific nutrient” and thus has extensive applications in the fields of pharmaceuticals and functional foods [3]. It has shown that PS has a positive impact on people’s emotions, cognition, and memory, and can significantly promote the growth of primary hippocampal nerve synapses, and has obvious effects on the prevention and alleviation of Alzheimer’s disease [4,5]. Recent studies have shown that PS on the outer leaflet of the cell membrane is closely related to acute inflammation and abnormal coagulation caused by COVID-19, which is of great significance for the pathophysiological study of COVID-19 [6]. With the in-depth research on the efficacy and application of PS, its market demand is growing. Traditionally, PS is mainly produced by extracting from animals and plants, but it requires a complex separation and extraction process with the generation of a large amount of organic wastewater, which makes it difficult to ensure the quality and safety of the product [7]. The preparation of PS by enzymatic modification of soybean phospholipids with PLD is more in line with the requirements of food and health products. Therefore, the enzymatic synthesis of PS mediated by PLD is a promising production method with great development potential, which has the advantages of simple operation, mild reaction conditions, and environmental protection.
At present, Bacillus cereus and Streptomyces are the main natural producers of PLD [8]. However, they are still faced with some problems, such as the long growth cycle, the complex cultural conditions and the low productivity, which limit the industrial production of PLD [9]. With the development of synthetic biology, it has been widely carried out by heterologous expression of functional enzymes in commonly used model chassis Escherichia coli [10]. Several studies have reported heterologously expressed PLD [11,12,13]. However, PLD is easily expressed in an insoluble form with a low enzyme activity. Wu et al. explored the influence of different plasmids on the heterologous expression of PLD from Streptomyces chromofuscus in E. coli [11]. It was found that when pET-28a was used as the expression vector, it could better promote the soluble expression of PLD in E. coli BL21(DE3). Combined with the optimization of the inducing and cultivation process, the activity of PLD was as high as 104.28 U/mL in the shake flasks. However, it was also shown that PLD was highly cytotoxic to E. coli, which caused some issues in the production of PLD, including plasmid instability, short-term PLD synthesis, celllysis and so on. The periplasmic space is a narrow space between the inner and outer membranes of E. coli. Compared with the intracellular environment, it has a more suitable oxidative environment for protein folding and a lower protease activity, which is conducive to the stability of recombinant proteins[14]. Xiong et al. used the gIIIC signal peptide to localize the recombinant PLD protein in the periplasmic space of E. coli, which indicated that celllysis did not occur and almost 99% PLD remained in the periplasmic space [15]. Moreover, they found that the expression of PLD led to the accumulation of phosphatidic acid and the enhancement of membrane permeability, which impacted the growth of E. coli. The addition of cations (Na+, K+, Li+, or Mg2+) could alleviate cell growth inhibition and significantly increase the synthesis of PLD. After the optimization of the fermentation process, the yield of PLD reached 1100 U/mL in the bioreactor, which is the highest reported intracellular accumulation of PLD in a cell factory at present [16].
Compared to the intracellular production method, the extracellular production of PLD has several distinct advantages that make it a promising alternative. First and foremost, extracellular production simplifies the downstream separation and purification process, as the recombinant protein is secreted outside the cell, avoiding the need for cell lysis and reducing the complexity of purification. It not only lowers the cost of purification but also saves time, making the overall process more efficient. Additionally, extracellular enzymes are often more stable and resistant to harsh environmental conditions, which can be beneficial for industrial applications. Furthermore, extracellular production allows for the efficient utilization of external substrates and promotes intercellular cooperation, which can be advantageous in certain biotechnological contexts [17,18]. Therefore, some studies focus on the secretory production of PLD. Hou et al. heterologously expressed the PLD gene from Streptomyces sp. in Bacillus subtilis, Pichia pastoris, and Corynebacterium glutamicum, respectively. It was found that the highest activity of PLD was observed in the engineered C. glutamicum, which reached 0.25 U/mL. Combined with the optimization of signal peptides, ribosome binding sites, and promoters, the activity of PLD was increased by 7.6 fold and was up to 1.9 U/mL [18]. Huang et al. heterologously expressed PLD from Streptomyces racemochromogenes in Bacillus subtilis. The activity of PLD was increased to 24.2 U/mL through signal peptide screening, plasmid and RBS optimization [19]. At present, there are few studies on the secretory production of PLD and the host is limited to Bacillus subtilis. More high-efficiency hosts with a clear genetic background and various genetic strategies need to be explored to improve the secretory production of PLD.
In this study, the commonly used E. coli was explored to secretively express PLD from Streptomyces antibioticus. Combining signal peptide screening and promoter optimization, the efficient secretory production of PLD was achieved in the engineered E. coli. On this basis, the effect of cell membrane permeability on the secreted production of PLD was explored via supplementing surfactants into the medium. Combined with the genetic and fermentation optimization, the extracellular PLD activity finally reached 33.26 U/mL in the bioreactor, which was the highest reported so far. Our work demonstrated the potential of E. coli in the secreted production of PLD and provided an alternative platform for the industrial production of PLD.
2. Materials and Methods
E. coli DH5α was used for plasmid construction and maintenance, while E. coli BL21(DE3) and E. coli TOP10 were used as the chassis to express PLD. All the strains used in this study were listed in Table S1. Luria-Bertani (LB) medium used for seed culture was composed of tryptone 10 g/L, yeast extract 5 g/L, and NaCl 10 g/L. Terrific Broth (TB) medium was used as fermentation medium for PLD production, which was composed of tryptone 12 g/L, yeast extract 24 g/L, glycerol 4 g/L, KH2PO4 23.1 g/L, K2HPO4 125.4 g/L. In this work, the optimized Terrific Broth (opTB) medium was composed of tryptone 12 g/L, yeast extract 24 g/L, glycerol 20 g/L, NaCl 23.4g/L, KH2PO4 23.1 g/L, K2HPO4 125.4 g/L.
2.1. Plasmid Construction
All the plasmids and primers used in this work are listed in Table S2 and Table S3. All restriction enzymes used in this work were purchased from NEB, while the PCR recovery kits, plasmid miniprep kits, and gel recovery kits were purchased from OMEGA. The PLD gene S. antibioticus were codon-optimized and synthesized by Genewiz, which was inserted into the vector pET28a at the NheI and BamHI or the vector pET22b containing the signal peptides PelB at the NdeI and HindIII sites. The signal peptides DsbA, OmpA, TorA, FhuD and PhoA were amplified from the genome of E. coli and assembled into the vector pET22b to replace the signal peptides PelB to explore their effects on the secretory production of PLD. Similarly, the PLD gene from S. antibioticus and OmpA signal peptide was assembled into the vector pBAD-gⅢC to explore the effect of promoter pBAD on the expression of PLD.
2.2. Strains Cultivation
The single colonies were inoculated into 4 mL LB medium supplemented with 0.1% (v/v) antibiotics (50 mg/mL kanamycin, 100 mg/mL ampicillin), which were then cultivated overnight in a rotary shaker at 37 °C, 200 rpm to finish the seed culture process. Afterward, 1 mL seed broth was inoculated into 250 mL flasks containing 50 mL TB medium supplemented with antibiotics and cultured at 37 °C, 200 rpm. When OD600 reached 6.0, the cells were obtained after centrifuged at 6000× g, −4 °C for 5 min. The cells were then transferred into 250 mL flasks containing 50 mL opTB medium supplemented with 0.2 mM IPTG or 0.2% (v/v) 200 mg/mL L-arabinose solution to induce the expression of PLD and cultured at 18 °C, 200 rpm for 48 h to finish the fermentation process.
2.3. Fed-Batch Fermentation
A 5-L bioreactor (Bailun BioTechnology Co., Shanghai, China) containing 3 L opTB medium was used to conduct the fed-batch fermentation. The strain was incubated in 500 mL flasks containing 100 mL TB medium and cultivated at 37 °C, 200 rpm for 6–7 h to complete seed cultivation. The two-stage cultivation strategy was employed in the bioreactor cultivation. In the first stage, 4% (v/v) inoculum was transferred into a bioreactor, which was then cultivated at 37 °C, 200 rpm, 4.0 vvm, pH 7.0 (controlled by 10% ammonia and 10% phosphoric acid) for 16 h with the OD600 about 18. Afterward, 0.2% (v/v) 200 mg/mL L-arabinose solution was added to the medium, and then the culturing conditions were changed to 18 °C, 200 rpm, 1 vvm to start the second PLD production stage.
2.4. Analytical Methods
2.4.1. Quantitative Real-Time PCR (qPCR)
Sample cells of strains ECPLD3 and ECPLD4 were harvested at 6 h to quantify the relative expression levels of the PLD gene without induction. Total RNA was extracted from the samples following the manufacturer’s instructions of the Total RNA Midi Kit (Omega BIO-TEK, Norcross, Georgia). Subsequently, cDNA was synthesized from the total RNA using the NovoScript® Plus All-in-one 1st Strand cDNA Synthesis SuperMix (gDNA Purge) Reverse Transcription Kit (Novoprotein Scientific Inc., Shanghai, China). The synthesized cDNA was then amplified in a qPCR for relative quantification of mRNA levels with the NovoStart® SYBR qPCR SuperMix Plus kit (Novoprotein Scientific Inc., Shanghai, China). The qPCR reactions were carried out using a LongGene Q2000A instrument, and the Ct value for each gene was obtained. GAPDH encoding glyceraldehyde-3-phosphate dehydrogenase was employed as the housekeeping gene, and the relative transcription levels of the PLD gene were calculated using the 2−ΔΔct method [20].
2.4.2. Analysis of PLD Activity
Cell samples were collected by centrifugation at 10,000× g for 5 min. The supernatant was used to determine the extracellular PLD activity. When measuring intracellular PLD activity, the cells were collected and resuspended in 20 mM Tris-HCl buffer (pH = 8.0). The resuspended cells were disrupted by sonication at 200 W for 15 min (3 s pulse and 7 s interval) in an ice-water bath using an Ultrasonic Cell Crusher, which was then centrifuged at 4 °C, 6000× g for 10 min to collect the supernatant for the determination of the intracellular PLD activity.
PLD activity was measured according to previous reports with minor modifications [21]. 1% Triton X-100 and 0.1 M CaCl2 were added to Tris-HCl solution (0.02 M, pH = 7.2) to prepare the substrate buffer, which was used to dissolve 0.7% (w/v) phosphatidylcholine to prepare the substrate solution. 0.1 M EDTA was added into 1 M Tris-HCl buffer with pH controlled to 8.0 to prepare the reaction termination solution. 0.15% phenol and 0.125% 4-aminoantipyrine were added to the Tris-HCl buffer (0.1 M, pH = 8) to make a color development solution. In the detection process, 400 μL substrate buffer and 100 μL substrate solution were mixed, while 10 μL diluted crude enzyme solution was added to start the reaction at 37 °C for 10 min. Subsequently, 200 μL reaction termination solution was immediately added and was boiled for 5 min to stop the reaction. After the samples were cooled down to room temperature, 100 μL color development solution, 10 μL of choline oxidase solution (25 U/mL), and 5 μL of peroxidase solution (10 U/mL) was added, which was then incubated at 37 °C for 3 h to measure the absorbance at 505 nm. A calibration curve was developed based on a series of standard choline chloride solutions. Under the above conditions, the amount of enzyme that can produce 1 μmol of choline per minute using PC as the substrate was defined as one PLD enzyme activity unit (U).
2.4.3. Detection of Plasmid Stability
The fermentation broth was diluted to 103 cfu/mL, which was spread on LB plates with and without antibiotics, respectively. The plates were incubated at 37 °C for 12 h, and the number of single colonies was counted. Plasmid stability was explored by calculating the ratio of the colonies’ number on the LB plate with antibiotics to without antibiotics [20].
2.4.4. Analysis of Cell Growth and Glycerol Consumption
The cell growth was detected by measuring the density at OD600 with a UV–vis spectrophotometer. The fermentation broth was centrifuged at 10,000× g for 10 min and the supernatant was prepared as samples for glycerol consumption using HPLC (Shimadzu, Kyoto, Japan) equipped with the refractive index detector (set at 50 °C). The analysis process was conducted using the Bio-Rad (Hercules, CA, USA) Aminex HPX-87H ion exchange column, which was eluted with 5 mM H2SO4 with a flow rate of 0.6 mL/min at a temperature of 35 °C. All the data were obtained from at least three independent experiments for statistical analysis, and their significance was calculated using GraphPad PRISM 9 software.
3. Results and Discussion
3.1. The Expression of PLD from S. antibioticus in E. coli
PLD from S. antibioticus was reported to have excellent transphosphatidylation activity and broad substrate specificity [22], which was assembled into the plasmid pET28a and introduced into the host E. coli BL21(DE3) to explore its expression. It was found that SaPLD achieved soluble expression without forming inclusion bodies in E. coli BL21(DE3). After 48 h fermentation cultivation, the intracellular enzyme activity of SaPLD was 4.3 U/mL, while the extracellular activity was 0.55 U/mL, which accounted for only 11.3% of the total enzyme activity. It is similar to the report that the codon-optimized PLD can be solubly expressed in E. coli, but most of the enzyme is stored intracellularly rather than secreted extracellularly [11]. PLD has a certain hydrolytic effect on the phospholipid bilayer of the cell membrane, which limits cell growth and the accumulation of PLD. Secreting PLD enzymes outside the cell can alleviate the cells’ toxicity, avoid degradation by endogenous proteases, and simplify subsequent separation and purification processes. Generally, the secretory production of the target protein can be achieved through the signal peptide positioned at the N/C-terminus of the protein, which directs the synthesized protein to be secreted outside the cells [23]. Consequently, suitable signal peptides were further screened in our work to facilitate the extracellular secretion of the PLD enzyme and enhance its synthesis efficiency.
3.2. Screening a Compatible Signal Peptide to Promote the Secretory Production of SaPLD
Different signal peptides possess varying secretion efficiencies for exogenous proteins. Although some studies have predicted the matching between signal peptides and different proteins, the specific relationship remains unclear [24]. Therefore, various common secretory signal peptides were carried out N-terminal fusions with SaPLD to explore their secretion efficiency in our work. The plasmid pET22b carrying the signal peptide pelB was widely used for the secretory production of heterologous proteins in E. coli [25], which was thus selected to replace the plasmid pET28a to express SaPLD. Five commonly used secretion signal peptides OmpA, FhuD, DsbA, PhoA, and TorA in E. coli were assembled into plasmid pET28b to replace the signal peptide pelB respectively to explore their effects on the secretory production of SaPLD.
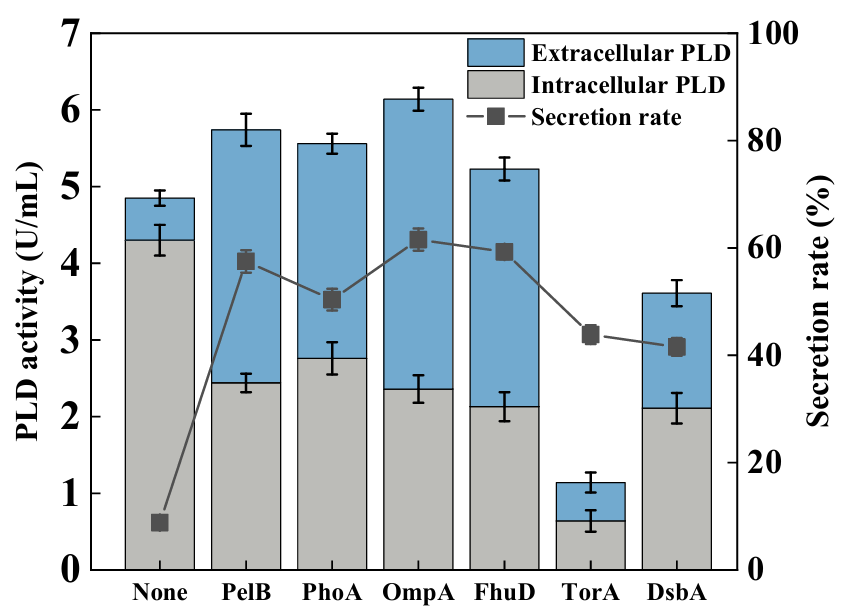
As shown in Figure 1, among the six signal peptides, PelB, PhoA, OmpA, and FhuD not only increased the extracellular enzyme activity but also improved the total enzyme activity. The PelB and PhoA signal peptides are derived from the pectate lyase of Erwinia carotovora and the alkaline phosphatase protein of E. coli, respectively. Both of them achieve the secreting functions through the SecB-dependent post-translational translocation mode and were commonly used to produce extracellular recombinant proteins in E. coli [26,27]. The OmpA signal peptide is derived from the outer membrane protein of E. coli and achieves post-translational translocation through the Sec system [28]. The FhuD and DsbA signal peptides are derived from a periplasmic transport protein, which is responsible for the transmembrane transport of ferric hydroxamate in E. coli. The fusion expression of OmpA and FhuD could promote the accumulation of target protein in the periplasmic space and enhance its solubility [29]. It was suspected that these four signal peptides could enable the target proteins to be accumulated in the periplasmic space or outside the cell in the form of unfolded polypeptide chains and then refolded into active proteins, which avoided the damage to the cells caused by PLD and thus led a higher total enzyme activity.
Although DsbA and TorA signal peptides are also derived from periplasmic transport proteins, the fusion expression resulted in a decrease in PLD activity. When the DsbA signal peptide was fused and expressed with PLD, the extracellular enzyme activity significantly increased but the total enzyme activity decreased. It was suspected that the DsbA signal peptide promoted the secretion of PLD protein, but the part of proteins may have failed to refold correctly during this process, which negatively affected the total enzyme activity. The TorA signal peptide is derived from a soluble periplasmic molybdenum enzyme, which is a classic signal peptide and was first discovered to be secreted through the Tat system in E. coli [30]. It did not rely on the Sec system and was capable of transporting fully folded proteins. But, the fusion expression of TorA signal peptide with PLD resulted in a decrease in both the extracellular enzyme activity and the total enzyme activity. It was speculated that the TorA signal peptide guided the complete folding of PLD, which was not conducive to its translocation to the outside of the cell.
Based on the above results, the OmpA signal peptide showed the best compatibility with PLD. The total enzyme activity reached 6.14 U/mL, with an increase of 26.6% compared to the activity without signal peptide. Meanwhile, the extracellular enzyme activity reached 3.78 U/mL, accounting for 61.6% of the total enzyme activity, which meant that the OmpA signal peptide could promote the expression of PLD and greatly improve the secretion of PLD outside the cells.
3.3. Replacing the Promoter to Enhance the Secretory Production of PLD
Promoters play a crucial role in the regulation of the transcriptional level. In the expression of recombinant proteins, it is necessary to select suitable promoters with different strengths and characteristics to achieve high-level expression [31]. Strong promoters are prone to cause leaky expression, which can generate a certain degree of transcription even without the addition of an inducer [32]. For proteins that are somewhat toxic to the host, even slight leaky expression will have a great impact on the yield [33]. Many studies have already proved that different promoters are of vital importance for the production of recombinant PLD in E. coli [15,18].
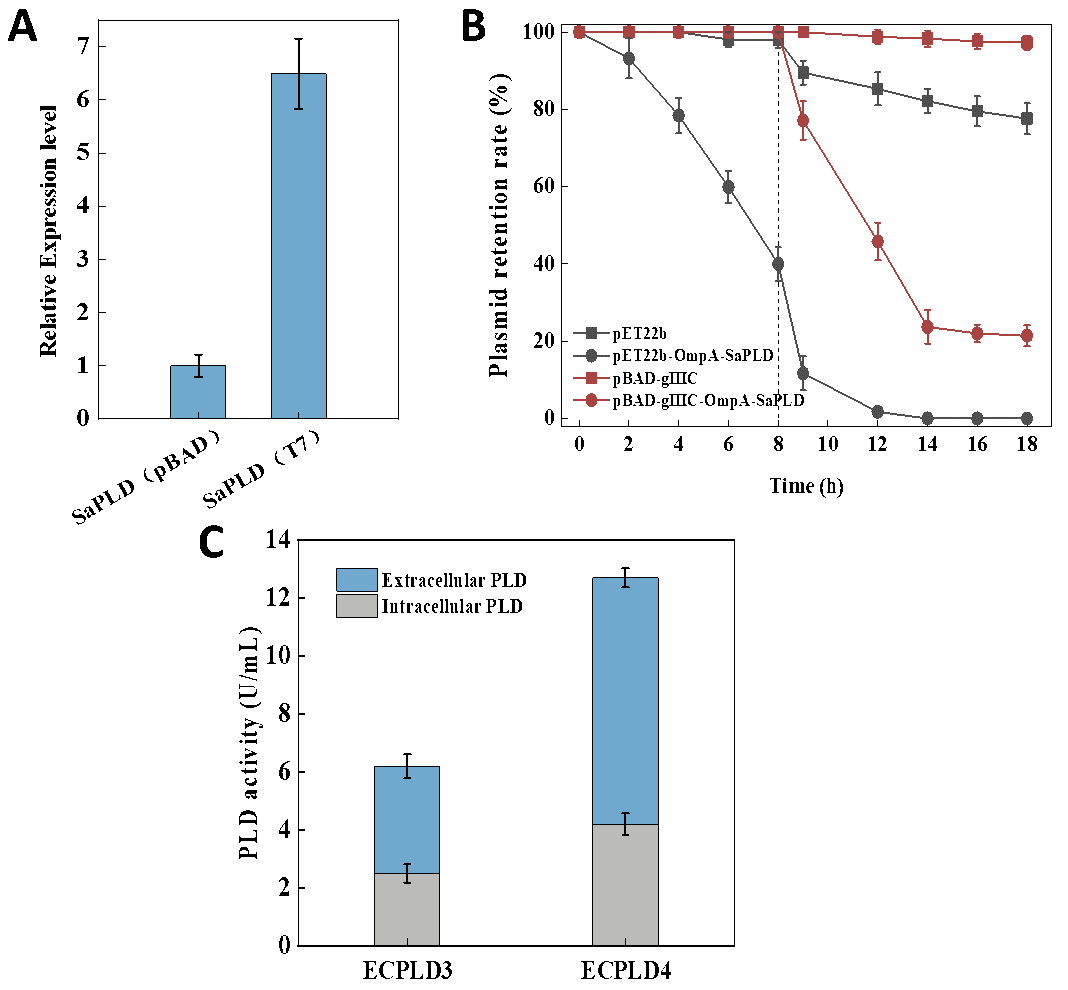
T7 promoter is a commonly used strong promoter in E. coli, which can be specifically recognized by T7 RNA polymerase and quickly initiate the transcription process, driving a very high level of transcription. However, the presence of other transcription factors that can bind to the T7 promoter as well as the high activity of T7 RNA polymerase in the host cells, lead to leaky expression [32]. The pBAD promoter is induced by the pentose sugar arabinose, which can bind to the regulatory protein AraC, thereby activating the pBAD promoter and initiating the transcription of downstream genes. In the absence of arabinose, the activity of the promoter is inhibited, which gives the target gene a relatively low level of leaky expression [34]. Considering the toxicity of PLD to cells, the pBAD-gⅢC plasmid containing the pBAD promoter was used to explore its effect on the expression of PLD. Firstly, the expression levels of PLD were compared under the control of T7 promoter and the pBAD promoter without inducer. As shown in Figure 2A, when the T7 promoter was used, the leakage expression level of SaPLD was 6.49 times higher than that of the pBAD promoter without the addition of the inducer. Although there was a lac operator system was contained in the plasmid pET22b to control the leakage expression, the basal expression of the T7 promoter was still not strictly inhibited according to the qPCR result. In addition, the peptone added in the TB medium may contain a trace amount of lactose, which was suspected to activate T7 promoter [35]. Compared with the T7 promoter, the pBAD promoter was stricter before the addition of the inducer.
The genetic stability of recombinant bacteria is determined by the holding rate of the recombinant plasmid. Plasmid stability is an important factor affecting protein expression and directly influences the yield of the target protein [36]. Therefore, the stabilities of plasmids pET22b and pBAD-gⅢC were explored during the expression of PLD in our work (Figure 2B). We compared the retention rate of plasmids pET22b, pBAD-gⅢC, pET22b-OmpA-SaPLD and pBAD-gⅢC-OmpA-SaPLD during the cell growth stage (0–8 h) and the PLD expression stage after adding inducer (8–18 h). In the cell growth stage, the empty plasmids pET22b and pBAD-gⅢC had preferable stabilities. Even 0.2 mM IPTG and 2% L-arabinose were added to induce expression, the plasmids pET22b and pBAD-gⅢC still exhibited a relatively high retention rate. Surprisingly, the plasmid pBAD-gⅢC-OmpA-SaPLD also showed considerable stability in the cell growth stage with a plasmid retention rate of 100%, which indicated that the low-level background expression of PLD driven by the pBAD promoter did not affect the stability of the plasmid. And there was still a 23.7% plasmid retention rate at the 14th hour of fermentation. In contrast, the strain carrying the plasmid pET22b-OmpA-SaPLD showed an obvious plasmid instability with a 40% retention rate in the cell growth stage, which was decreased to 1.7% after 12 h fermentation with the addition of the inducer. These results indicated that the pBAD-gⅢC plasmid was more stable within cells.
Therefore, the intracellular and extracellular enzyme activities of PLD were measured and compared after the cultivation of the stains for 48 h (Figure 2C). The extracellular enzyme activity of the strain ECPLD3 carrying plasmid pET22b-OmpA-SaPLD was 3.7 U/mL, with the total enzyme activity reaching 6.2 U/mL, while the counterparts in the strain ECPLD4 carrying plasmid pBAD-gⅢC-OmpA-SaPLD were 8.5 U/mL and 12.7 U/mL, which were about 2.3 and 2.1 times higher than that in strain ECPLD3, respectively. This indicated that the strictly regulated promoter pBAD with a lower leaky expression level was more conducive to the expression of the slightly toxic protein than T7 promoter, which significantly accelerated the production of PLD in E. coli.
3.4. Regulation of Cell Permeability to Promote the Accumulation of PLD
In order to enhance the secretory production of PLD, the OmpA signal peptide was fused at the N-terminus of SaPLD, and approximately 60% of PLD was secreted extracellularly. But there was still about 40% PLD remained intracellular and failed to be secreted. It was suspected that the ability of the Sec system was limited for the secretory production of the recombinant proteins due to the complex membrane structure of E. coli [37]. Signal peptides could usually guide proteins to cross the inner membrane and enter the periplasmic space, but it was rather difficult for proteins to be secreted across the cell wall from the periplasmic space. Therefore, regulation of the permeability of cell walls was another available way to promote the extracellular secretion of recombinant proteins out of cells [38].
The genes mrcB and dacB encode penicillin-binding proteins PBP1b (high molecular weight) and PBP4 (low molecular weight) in E. coli, which play a crucial role in the formation of peptidoglycan linkage and peptidoglycan network cross-linking during the synthesis process of the peptidoglycan layer in the cell wall [39]. Knocking out the mrcB and dacB could inhibit the synthesis of cell wall peptidoglycan, thereby improving cell permeability and significantly enhancing the production of extracellular recombinant proteins [40]. Herein, they were also knocked out to explore their effects on cell growth and PLD production. However, it was found that the knockout of two genes obviously inhibited cell growth and the production of PLD (Figure S1). It might be because the deletion of mrcB and dacB destroyed the peptidoglycan network in the cell wall and negatively affected the protectiveness of cells [41]. Therefore, knocking out mrcB and dacB was detrimental to the production of PLD.
Surfactant molecules containing hydrophilic and hydrophobic groups can interact with the hydrophobic parts of the phospholipid bilayer of the cell membrane, which leads to the loose arrangement of the phospholipid molecules and increases the fluidity of the cell membrane [42]. As the concentration of surfactants increases, they will form structures such as micelles on the cell membrane, further destroying the integrity of the cell membrane and resulting in the appearance of some small holes or channels on the cell membrane [43]. This enables some substances freely to pass through the cell membrane. Therefore, we also explored the impact of surfactants on the secretion of PLD in this work.
Different surfactants have different mechanisms on cells. Ca2+ can reduce the hydrolysis of peptidoglycan by peptidoglycan hydrolase and stabilize the lipopolysaccharide in the cell wall, which may help alleviate the lytic effect of toxic substances on cells [44]. Glycine can be incorporated into peptidoglycan precursors and modifies peptidoglycan, which is usually considered to increase cell membrane permeability [45]. SDS is an anionic surfactant that can inhibit the biosynthesis of lipase in the cell membrane and results in insufficient synthesis of membrane phospholipids to increase the cell membrane’s permeability [46]. Tween 80 and Tween 20 are a kind of amphiphilic non-ionic surfactants in the polysorbate family, which can improve the fluidity of the cell membrane by changing the composition of fatty acids [47]. In addition, Tween 80 and Tween 20 can also stabilize proteins through interfacial competition and are often used as protein protectors. Triton X-100 is a non-ionic surfactant that can destroy the lipid structure and interfere with cell membrane lipoproteins to promote the secretion of intracellular substances [48]. PEG4000 can disperse the phospholipid molecules in the lipid bilayer and promote structural rearrangement which is often used to mediate cell fusion and is also considered helpful for releasing intracellular proteins [49]. Therefore, 0.1% (w/v) calcium chloride, glycine, SDS, Tween 20, Tween 80, Triton X-100 and PEG4000 were prepared as surfactants, respectively and added in the medium during the induction stage of ECPLD4 strain to explore their impacts on the secretory production of PLD (Figure 3A and Figure S2).
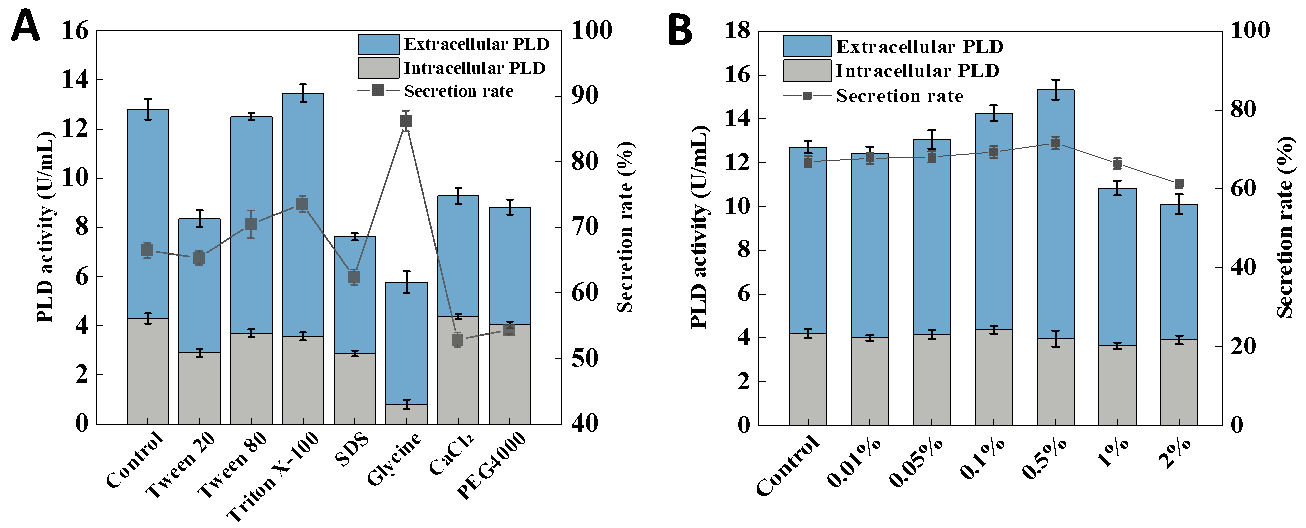
Different surfactants showed different effects on the secretory production of PLD. The addition of calcium chloride, glycine, SDS, Tween 20 and PEG4000 led to the reduction in both total activity and extracellular activity of PLD. Surprisingly, the highest secretion rate was obtained, with 0.1% (w/v) glycine being added, reaching 86.1%. However, the total enzyme activity of PLD was relatively low due to a significant decrease in the cell growth of ECPLD4. It was reported that glycine was a competitive analogue of L-alanine or D-alanine and led to the accumulation of uridine-N-acetylmuramic acid pentapeptide, which affected the synthesis of peptidoglycan for the cell wall [50]. Zou et al. also observed a similar phenomenon in the production of pullulanase by the engineered E. coli [51]. Namely, glycine could significantly improve pullulanase’s secretion efficiency but inhibit cell growth.
Among these surfactants, Tween 80 and Triton X-100 could significantly promote the extracellular secretion of PLD. The addition of 0.1% (w/v) Tween 80 led to the extracellular PLD activity reaching 8.8 U/mL with the total activity of 12.5 U/mL and the extracellular secretion rate of 70.40%, while 0.1% (w/v) Triton X-100 led to the extracellular enzyme activity up to 9.89 U/mL with the total activity of 13.46 U/mL and the extracellular secretion rate of 73.47%. Therefore, Triton X-100 was considered the optimum surfactant for the production of PLD by E. coli.
In order to know the effect of Triton X-100 in detail, different concentrations (0.01%, 0.05%, 0.1%, 0.5%, 1% and 2%) of Triton X-100 were added to the medium, and the extracellular PLD enzyme activities were detected and compared after cultivation for 48 h (Figure 3B). The extracellular PLD activity gradually improved as the addition amount of Triton X-100 increased. When 0.5% (w/v) Triton X-100 was added, the PLD activity reached the highest and the extracellular secretion rate reached 74.17%. However, when the additional amount of Triton X-100 continued to increase, the extracellular enzyme activity decreased significantly. It might be that the excessive concentration of Triton X-100 damaged the cell membrane of E. coli and had an adverse impact on PLD production. Yang et al. also reached a similar conclusion that Triton X-100 as a surfactant and PLD as a cytotoxic protein both had a negative impact on cell growth. However, a good balance between them could effectively promote the extracellular release of PLD [13].
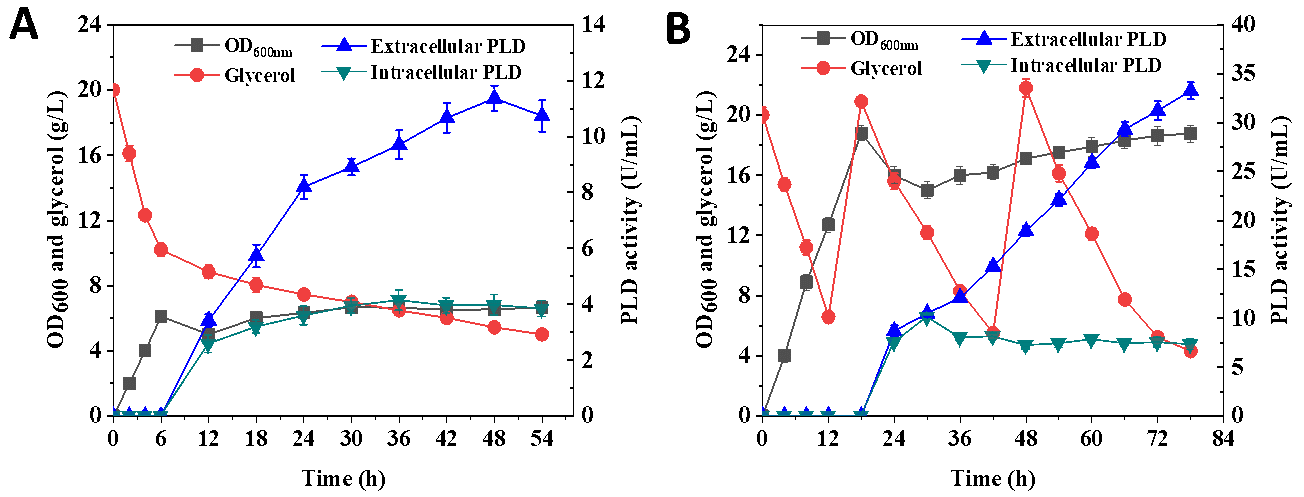
Based on the above conditions, the production of PLD by the engineered strain ECPLD4 was investigated in detail in shake flasks (Figure 4A). After being inoculated into the fermentation medium following seed cultivation, the strain grew well in the early stage and the OD600 gradually increased, which reached 6.11 at 6 h of cultivation. Subsequently, the cultivation temperature was controlled at 18 °C, and arabinose was added to induce the synthesis of PLD. In the enzyme production stage, cell growth was limited and the OD600 remained at around 6.6, while the carbon source glycerol also remained at 5.44 g/L after 48 h of cultivation, which was suspected to own to the negative effect of PLD synthesis on the cell growth. The accumulation of PLD gradually increased, and the extracellular PLD activity reached the highest level of 11.37 U/mL at 48 h with a total activity of 15.33 U/mL (Figure 4A), which was about 1.34 and 1.21 times higher than that of the control group without addition of surfactants. It demonstrated that the co-optimization of genetic and fermentation processes significantly improved the capability for extracellular synthesis of PLD by the engineered E. coli.
3.5. PLD Production in Fed-Batch Fermentation by ECPLD4 Strain
To further increase the yield of PLD, the fed-batch fermentation was carried out in a 5L reactor. Similar to shake flask cultivation, the fed-batch fermentation process was also divided into the cell growth stage and the PLD synthesis stage. As shown in Figure 4B, the OD600 increased rapidly during the cell growth stage and reached 18.8 at 18 h. Afterward, the inducer arabinose and 60 g glycerol were added to the medium to start the PLD synthesis stage. The activity of PLD was gradually increased with the consumption of glycerol. Subsequently, 60 g glycerol was supplemented at the incubation of 48 h to improve further the accumulation of PLD (Figure 4B). The intracellular PLD activity reached a maximum of 10.1 U/mL at 30 h, while the extracellular enzyme activity gradually increased and reached 33.25 U/mL at the incubation of 78 h. Currently, there are few studies on the extracellular production of PLD, among which B. subtilis showed the preferable extracellular yield of PLD (24.2 U/mL) [19]. In our work, the extracellular PLD activity reached the highest level reported so far using a certain amount of glycerol as the substrate by the engineered strain ECPLD4. It was demonstrated that the engineered E. coli has promising potential for extracellular synthesis of PLD. Moreover, glycerol was a by-product of biodiesel, making PLD production more economically feasible. Our work laid a good foundation for the industrial production of PLD by the microbial cell factories from the low-cost feedstock.
Figure 1. Comparison the effects of different signal peptides on the secretory production of PLD in <i>E. coli</i>.
Figure 2. The effect of promoters on the secretory production of PLD. (<b>A</b>): relative expression level (in comparison to the expression leve of housekeeping gene) of SaPLD controlled by pBAD promoter and T7 promoter without inducer; (<b>B</b>): comparison of the stabilities of pBAD-gⅢC plasmid and pET22b plasmid for the expression of SaPLD; (<b>C</b>): comparison of intracellular and extracellular PLD activities produced by strains ECPLD3 and ECPLD4.
Figure 3. Effect of surfactants on the secretary expression of PLD by ECPLD4. (<b>A</b>): comparison of different surfactants on PLD production; (<b>B</b>): effect of different concentrations of Triton X-100 on PLD production.
Figure 4. Detail exploring PLD production by the engineered strain ECPLD4 in shake flasks and fed-batch fermentation process. (<b>A</b>): Fermentation profile of glycerol consumption, extracellular and intracellular PLD activity, OD<sub>600</sub> for strain ECPLD4 in shake flasks cultivation; (<b>B</b>): Fermentation profile of glycerol consumption, extracellular and intracellular PLD activity, OD<sub>600</sub> for strain ECPLD4 in fed-batch fermentation process.
4. Conclusions
In this study, E. coli served as a chassis to explore efficient heterologous expression and secretion of PLD. N-terminal fusion secretion signal peptides were explored to enhance PLD secretion. Among six signal peptides, the OmpA signal peptide proved the most effective, increasing extracellular PLD activity to 3.78 U/mL with a total activity of 6.14 U/mL. On this basis, the effects of plasmids and promoters on the expression of PLD were investigated. The plasmid pBAD-gⅢC with pBAD promoter was identified as the most suitable, resulting in a total PLD activity of up to 12.7 U/mL with an extracellular enzyme activity of 8.5 U/mL. Furthermore, the cellular membrane permeability was regulated to improve the secretory production of PLD by manipulating the key protein expression in the cell membrane and the addition of surfactants. The addition of 0.5% (w/v) Triton X-100 was proven to be more effective, which improved the total PLD activity to 15.33 U/mL and the extracellular activity to 11.37 U/mL with a secretion rate of 74.17%. Finally, fed-batch fermentation was conducted in a 5-L fermentor, and the maximum extracellular PLD activity achieved 33.25 U/mL after being cultivated for 78 h, which was the highest level reported so far. Our work demonstrated the effectiveness of genetic and process engineering strategies for the expression and extracellular secretion of PLD in E. coli, which helped to overcome the insoluble expression and inadequate secretion of PLD and laid a good foundation for its industrial production.
Supplementary Materials
The following supporting information can be found at: https://www.sciepublish.com/article/pii/494, Table S1: Strains used in this study; Table S2: Plasmids used in this study; Table S3: Primers and synthetic oligos used in this study; Figure S1. PLD production, cell growth and glycerol consumption by the engineered ECPLD4 after knockout of mrcB and dacB; Figure S2. Comparison of the effects of different surfactants with 0.1% and 0.5% addition level on PLD production.
Author Contributions
H.L.: Conceptualization, Data curation; H.L. and Y.Y. Writing-original draft; Y.Y., T.W. and Y.N.: Investigation, Methodology; L.D. and F.W.: Writing-review & editing, Supervision, Funding acquisition, Project administration.
Ethics Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
This work was funded by the National Key Research and Development Program of China (2022YFC2106100), the National Natural Science Foundation of China (22208011, 20308020).
Declaration of Competing Interest
The authors declared that they have no conflicts of interest to this work.
References
1.
Zhang Z, Chen M, Xu W, Zhang W, Zhang T, Guang C, et al. Microbial phospholipase D: Identification, modification and application. Trends Food Sci. Tech. 2020, 96, 145–156. [Google Scholar]
2.
Zhang P, Gong J-S, Qin J, Li H, Hou H-J, Zhang X-M, et al. Phospholipids (PLs) know-how: Exploring and exploiting phospholipase D for its industrial dissemination. Crit. Rev. Biotechnol. 2021, 41, 1257–1278. [Google Scholar]
3.
Chaung H-C, Chang C-D, Chen P-H, Chang C-J, Liu S-H, Chen C-C. Docosahexaenoic acid and phosphatidylserine improves the antioxidant activities in vitro and in vivo and cognitive functions of the developing brain. Food Chem. 2013, 138, 342–347. [Google Scholar]
4.
Moré MI, Freitas U, Rutenberg D. Positive effects of soy lecithin-derived phosphatidylserine plus phosphatidic acid on memory, cognition, daily functioning, and mood in elderly patients with Alzheimer’s disease and dementia. Adv. Ther. 2014, 31, 1247–1262. [Google Scholar]
5.
Bie N, Li J, Li C, Lian R, Qin L, Wang C. Protective effect and mechanism of docosahexaenoic acid on the cognitive function in female APP/PS1 mice. Food Funct. 2021, 12, 11435–11448. [Google Scholar]
6.
Argañaraz GA, Palmeira JdF, Argañaraz ER. Phosphatidylserine inside out: A possible underlying mechanism in the inflammation and coagulation abnormalities of COVID-19. Cell Commun. Signal 2020, 18, 190. [Google Scholar]
7.
Zhou WB, Gong JS, Hou HJ, Li H, Lu ZM, Xu HY, et al. Mining of a phospholipase D and its application in enzymatic preparation of phosphatidylserine. Bioengineered 2018, 9, 80–89. doi:10.1080/21655979.2017.1308992. [Google Scholar]
8.
Nakazawa Y, Sagane Y, Sakurai S, Uchino M, Sato H, Toeda K, et al. Large-scale production of phospholipase D from Streptomyces racemochromogenes and its application to soybean lecithin modification. Appl. Biochem. Biotechnol. 2011, 165, 1494–1506. doi:10.1007/s12010-011-9370-4. [Google Scholar]
9.
Zhao Y, Xu Y, Yu F, Zhang C. Identification of a novel phospholipase D gene and effects of carbon sources on its expression in Bacillus cereus ZY12. J. Microbiol. 2018, 56, 264–271. doi:10.1007/s12275-018-7529-1. [Google Scholar]
10.
Zhang H, Li X, Liu Q, Sun J, Secundo F, Mao X. Construction of a super-folder fluorescent protein-guided secretory expression system for the production of phospholipase D in Bacillus subtilis. J. Agric. Food Chem. 2021, 69, 6842–6849. doi:10.1021/acs.jafc.1c02089. [Google Scholar]
11.
Wu R, Cao J, Liu F, Yang M, Su E. High-level soluble expression of phospholipase D from Streptomyces chromofuscus in Escherichia coli by combinatorial optimization. Electron. J. Biotechnol. 2021, 50, 1–9. [Google Scholar]
12.
Zhang H, Chu W, Sun J, Liu Z, Huang WC, Xue C, et al. Combining cell surface display and DNA-shuffling technology for directed evolution of Streptomyces phospholipase D and synthesis of phosphatidylserine. J. Agric. Food Chem. 2019, 67, 13119–13126. [Google Scholar]
13.
Yang L, Xu Y, Chen Y, Ying H. Efficient extracellular expression of phospholipase D in Escherichia coli with an optimized signal peptide. IOP Conf. Ser. Mater. Sci. Eng. 2018, 301, 012105. [Google Scholar]
14.
Kaur J, Kumar A, Kaur J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. doi:10.1016/j.ijbiomac.2017.08.080. [Google Scholar]
15.
Xiong W, Zeng X, Ho SH, Ling X, Shen L, Yao C, et al. Strategies for achieving high-level and stable production of toxic Streptomyces phospholipase D in Escherichia coli. J. Chem. Technol. Biotechnol. 2019, 94, 1220–1229. [Google Scholar]
16.
Xiong W, Luo W, Zhang X, Pan X, Zeng X, Yao C, et al. High expression of toxic Streptomyces phospholipase D in Escherichia coli under salt stress and its mechanism. AIChE J. 2020, 66, e16856. [Google Scholar]
17.
Chen S, Xiong W, Zhao X, Luo W, Yan X, Lu Y, et al. Study on the mechanism of efficient extracellular expression of toxic Streptomyces phospholipase D in Brevibacillus choshinensis under Mg2+ stress. Microb. Cell Fact. 2022, 21, 41. [Google Scholar]
18.
Hou H-J, Gong J-S, Dong Y-X, Qin J, Li H, Li H, et al. Phospholipase D engineering for improving the biocatalytic synthesis of phosphatidylserine. Bioprocess Biosyst. Eng. 2019, 42, 1185–1194. [Google Scholar]
19.
Huang T, Lv X, Li J, Shin H-d, Du G, Liu L. Combinatorial fine-tuning of phospholipase D expression by Bacillus subtilis WB600 for the production of phosphatidylserine. J. Microbiol. Biotechnol. 2018, 28, 2046–2056. [Google Scholar]
20.
Liu H, Liu S, Ning Y, Zhang R, Deng L, Wang F. Metabolic engineering of Escherichia coli for efficient production of 1,4-butanediol from crude glycerol. J. Environ. Chem. Eng. 2024, 12, 111660. doi:10.1016/j.jece.2023.111660. [Google Scholar]
21.
Zhang P, Gong JS, Xie ZH, Su C, Zhang XM, Rao ZM, et al. Efficient secretory expression of phospholipase D for the high-yield production of phosphatidylserine and phospholipid derivates from soybean lecithin. Syn. Syst. Biotechnol. 2023, 8, 273–280. doi:10.1016/j.synbio.2023.03.006. [Google Scholar]
22.
Yamaguchi R, Akter S, Kanehama A, Iwamoto T, Hasegawa M, Ito A, et al. Improvement of solubility of phospholipase D from Streptomyces antibioticus in recombinant Escherichia coli and its application for the enzymatic synthesis of a non-natural plasmalogen. Lett. Appl. Microbiol. 2023, 76, ovad049. [Google Scholar]
23.
Güler-Gane G, Kidd S, Sridharan S, Vaughan TJ, Wilkinson TC, Tigue NJ. Overcoming the refractory expression of secreted recombinant proteins in mammalian cells through modification of the signal peptide and adjacent amino acids. PloS ONE 2016, 11, e0155340. [Google Scholar]
24.
Grasso S, Dabene V, Hendriks MM, Zwartjens P, Pellaux R, Held M, et al. Signal peptide efficiency: From high-throughput data to prediction and explanation. ACS Synth. Biol. 2023, 12, 390–404. [Google Scholar]
25.
Rahmatabadi SS, Askari S, Khademi F, Soleymani B. The study of different signal peptides in improvement of recombinant proteins solubility in E. coli: A review article. Curr. Proteom. 2024, 21, 129–139. [Google Scholar]
26.
Shi L, Liu H, Gao S, Weng Y, Zhu L. Enhanced extracellular production of is PETase in Escherichia coli via engineering of the pelB signal peptide. J. Agric. Food Chem. 2021, 69, 2245–2252. [Google Scholar]
27.
Liu W, Zhang R, Tian N, Xu X, Cao Y, Xian M, et al. Utilization of alkaline phosphatase PhoA in the bioproduction of geraniol by metabolically engineered Escherichia coli. Bioengineered 2015, 6, 288–293. [Google Scholar]
28.
Nielsen DW, Ricker N, Barbieri NL, Allen HK, Nolan LK, Logue CM. Outer membrane protein A (OmpA) of extraintestinal pathogenic Escherichia coli. BMC Res. Notes 2020, 13, 51. [Google Scholar]
29.
Zhang F, Fan X, Xu K, Wang S, Shi S, Yi L, et al. Development of a bacterial FhuD-Lysozyme-SsrA mediated Autolytic (FLSA) system for effective release of intracellular products. ACS Synth. Biol. 2022, 12, 196–202. [Google Scholar]
30.
Bageshwar UK, DattaGupta A, Musser SM. Influence of the TorD signal peptide chaperone on Tat-dependent protein translocation. PloS ONE 2021, 16, e0256715. [Google Scholar]
31.
Pouresmaeil M, Azizi-Dargahlou S. Factors involved in heterologous expression of proteins in E. coli host. Arch. Microbiol. 2023, 205, 212. [Google Scholar]
32.
Schuster LA, Reisch CR. Plasmids for controlled and tunable high-level expression in E. coli. Appl. Environ. Microb. 2022, 88, e00939-00922. [Google Scholar]
33.
Huleani S, Roberts MR, Beales L, Papaioannou EH. Escherichia coli as an antibody expression host for the production of diagnostic proteins: significance and expression. Crit. Rev. Biotechnol. 2022, 42, 756–773. [Google Scholar]
34.
Széliová D, Krahulec J, Šafránek M, Lišková V, Turňa J. Modulation of heterologous expression from PBAD promoter in Escherichia coli production strains. J. Biotechnol. 2016, 236, 1–9. [Google Scholar]
35.
Studier FW. T7 expression systems for inducible production of proteins from cloned genes in E. coli. Curr. Protoc. Mol. Biol. 2018, 124, e63. [Google Scholar]
36.
Nag N, Khan H, Tripathi T. Strategies to improve the expression and solubility of recombinant proteins in E. coli. In Advances in Protein Molecular and Structural Biology Methods; Elsevier: Amsterdam, The Netherlands, 2022; pp. 1– 12.
37.
Yoon S, Seo KS, Park N, Kim C, Dey P, Thornton JA, et al. Development of high-performance inducible and secretory expression vector and host system for enhanced recombinant protein production. Sci. Rep. 2024, 14, 30780. [Google Scholar]
38.
Maphosa S, Moleleki LN, Motaung TE. Bacterial secretion system functions: evidence of interactions and downstream implications. Microbiology 2023, 169, 001326. [Google Scholar]
39.
Thulin E, Andersson DI. Upregulation of PBP1B and LpoB in cysB mutants confers mecillinam (amdinocillin) resistance in Escherichia coli. Antimicrob. Agents Chemother. 2019, 63, e00612-19. doi:10.1128/AAC.00612-19. [Google Scholar]
40.
Yadav AK, Espaillat A, Cava F. Bacterial strategies to preserve cell wall integrity against environmental threats. Front. Microbiol. 2018, 9, 2064. [Google Scholar]
41.
Yang H, Wang F, Wang H, Lu X, Shen W, Chen X. Deleting mrdA and mrcB to significantly improve extracellular recombinant protein production in Escherichia coli. Biochem. Eng. J. 2019, 143, 185–195. [Google Scholar]
42.
Balantič K, Weiss VU, Allmaier G, Kramar P. Calcium ion effect on phospholipid bilayers as cell membrane analogues. Bioelectrochemistry 2022, 143, 107988. [Google Scholar]
43.
Schreier S, Malheiros SV, de Paula E. Surface active drugs: self-association and interaction with membranes and surfactants. Physicochemical and biological aspects. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2000, 1508, 210–234. [Google Scholar]
44.
Fitri Kusuma SA, Parwati I, Rostinawati T, Rukayadi Y, Subroto T. Improvement of extracellular secretion efficiency of recombinant proteins from Escherichia coli: Signal peptide fusion, surfactants addition, and phospholipase C coexpression. Drug Invent. Today 2019, 11, 2200. [Google Scholar]
45.
Egan AJ, Biboy J, van’t Veer I, Breukink E, Vollmer W. Activities and regulation of peptidoglycan synthases. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150031. [Google Scholar]
46.
Anosov A, Smirnova EY, Korepanova E, Shogenov I. The effects of SDS at subsolubilizing concentrations on the planar lipid bilayer permeability: Two kinds of current fluctuations. Chem. Phys. Lipids 2019, 218, 10–15. [Google Scholar]
47.
Nascimento TP, Sales AE, Porto TS, Costa RMPB, Breydo L, Uversky VN, et al. Purification, biochemical, and structural characterization of a novel fibrinolytic enzyme from Mucor subtilissimus UCP 1262. Bioprocess Biosyst. Eng. 2017, 40, 1209–1219. [Google Scholar]
48.
Romano S, Nele V, Campani V, De Rosa G, Cinti S. A comprehensive guide to extract information from extracellular vesicles: a tutorial review towards novel analytical developments. Anal. Chim. Acta 2024, 1302, 342473. [Google Scholar]
49.
Shah MKA, Azad AK, Nawaz A, Ullah S, Latif MS, Rahman H, et al. Formulation development, characterization and antifungal evaluation of chitosan NPs for topical delivery of voriconazole in vitro and ex vivo. Polymers 2021, 14, 135. [Google Scholar]
50.
Sidiq KR. Cell wall metabolism in Bacillus subtilis. Doctoral Dissertation, Newcastle University, Newcastle upon Tyne, UK, 2016.
51.
Duan X, Zou C, Wu J. Triton X-100 enhances the solubility and secretion ratio of aggregation-prone pullulanase produced in Escherichia coli. Bioresour. Technol. 2015, 194, 137–143. [Google Scholar]