Part of Special Issue:
Synthetic Biology in the Manufacturing of Chemicals and Fuels
Contents
Metabolic Engineering and Genome-Wide Adaptive Evolution for Efficient Reduction of Glycerol in Industrial Saccharomyces cerevisiae
Author Information
Other Information
1
The Hubei Provincial Key Laboratory of Yeast Function, Yichang 443003, China
2
National Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan 430070, China
*
Authors to whom correspondence should be addressed.
Received: 10 February 2025 Accepted: 13 March 2025 Published: 20 March 2025
© 2025 The authors. This is an open access article under the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/).
Synth. Biol. Eng.
2025,
3(1), 10004;
DOI: 10.70322/sbe.2025.10004
ABSTRACT:
The
production of glycerol as a major by-product during yeast-based bioethanol
fermentation arises directly from the need to re-oxidize excess NADH, which
reduces conversion efficiency. In this study, an
optimized Cas9-based genome editing
method was performed to develop a mixotrophic CO2-fixing industrial Saccharomyces cerevisiae by heterologous
expression of ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO form Pseudomonas sp.)
and phosphoribulokinase (PRK form Spinach). Additionally, the gene
encoding alcohol dehydrogenase (ADH2) responsible for converting ethanol to
acetaldehyde was deleted, while the great wall-family protein kinase Rim15 gene was overexpressed to facilitate the
reduction in glycerol content. The resulting CO2-fixing yeast M-2
led to a 21.5% reduction of the by-product glycerol in corn mash fermentation
cultures at 39 ℃. Moreover, we established a novel gene mutators mediated genome-wide mutations system that accumulates distinct
mutations in the industrial S. cerevisiae strains under the stress conditions to improve the robustness in the S. cerevisiae strains efficiently.
Keywords:
Saccharomyces cerevisiae; Gene
editing;
Glycerol yield;
Genome-wide mutation; Adaptive evolution; Robustness
1. Introduction
Ethanol, predominantly manufactured by the yeast Saccharomyces cerevisiae, has extensive application in various industries, including chemicals, beverages, bioethanol, pharmaceuticals, and cosmetics. It has been estimated that up to 4% of the sugar feedstock is converted into glycerol in the fermentation process of ethanol production by S. cerevisiae [1]. Although glycerol plays a physiological role in osmoregulation and redox balance, excessive production of glycerol can reduce the efficiency of sugar utilization, ultimately influencing the rate of ethanol production [2,3]. Bro et al. [4] overexpressed non-phosphorylating NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase(GAPN) from Streptococcus mutants to replace the NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase in S. cerevisiae, which the glycerol yield decreased by 40% and the ethanol yield increased by 3%.
In anaerobic cultures of S. cerevisiae that produce ethanol, the excess NADH produced during biosynthetic processes, including NAD+-dependent oxidative decarboxylations involved in the synthesis of acetyl-CoA and 2-oxoglutarate precursors, is re-oxidized by reducing part of the sugar substrate to glycerol [5]. Currently, utilizing CO2 as an electron acceptor for the reoxidation of NADH is a highly attractive metabolic engineering strategy, especially when CO2 reduction can be synchronized with the formation of the desired product [6]. Among the six naturally existing CO2 fixation pathways, the Calvin-Benson-Bassham(CBB) cycle is the most widely exploited for developing CO2-fixing S. cerevisiae strains [7,8]. Previous studies have demonstrated that certain strains of S. cerevisiae are capable of utilizing CO2 following the heterologous expression of the CBB cycle-associated enzymes, specifically ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and phosphoribokinase (PRK) [9,10]. Additionally, the incorporation of numerous replicates of a bacterial RubisCO expression cassette within S. cerevisiae, along with the GroEL/GroES chaperonin system from E. coli and the anaerobic inducible DAN1 promoter regulating PRK expression, results in an engineered S. cerevisiae strain that exhibits a 31% reduction in glycerol production [11]. When the alcohol dehydrogenase II (ADH2) gene was deleted in genetically modified S. cerevisiae, the resulting ethanol content increased by 18.58% with the decrease of glycerol, acetic acid, and lactic acid contents by 22.32%, 8.87%, and 16.82%, respectively [12]. Meanwhile, glycerol is primarily exported across the plasma membrane via the protein channel Fps1, which is regulated by extracellular osmolarity [13]. Fps1 is a member of the major intrinsic protein (MIP) family of channel proteins and contains six transmembrane domains [12]. The ∆FPS1 mutant of S. cerevisiae exhibits intracellular accumulation of glycerol, and the accumulation of glycerol further triggers other regulatory systems to reduce glycerol biosynthesis, leading to an increase in ethanol production [14].
Due to the complexity of metabolic and regulatory networks in microbial systems, obtaining phenotypes with good robustness through rational design is challenging. Currently, genome-scale evolution strategies such as Synthetic Chromosome Rearrangement and Modification by LoxP-mediated Evolution (SCRaMbLE) [15] in synthetic S. cerevisiae, Oligonucleotide-mediated Genome Engineering (YOGE and eMAGE) [16,17], and CRISPR-based multi-loci editing (CHAnGE, MAGESTIC, Target-AID, and yEvolvR) [18,19,20] have been extensively studied for improving the robustness of the strain. Genetic engineering via the CRISPR system is relatively straightforward in S. cerevisiae due to its excellent homologous recombination capability. However, industrial yeast strains are genetically more complex and often polyploid, making them more difficult to genetically manipulate than haploid laboratory strains [21]. To efficiently and rapidly manipulate the genome of industrial S. cerevisiae, Stovicek et al. [22] constructed the multi-copy 2μ plasmids to expressed S. pyogenes Cas9 protein and the corresponding gRNA, along with 90 bp double-stranded DNA as a repair template, and successfully achieved gene knockout with an efficiency of 65% to 78%. Lian et al. [23] successfully constructed higher-copy gRNA plasmids by optimizing the length of the resistance marker promoter and achieved a 100% efficiency in one-step knockout of four genes in diploid and triploid yeast.
In this study, we first optimized the CRISPR editing method in industrial S. cerevisiae. Subsequently, we developed a mixotrophic CO2-fixing industrial S. cerevisiae by heterologous expression of ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) and phosphoribulokinase (PRK) and modified endogenous metabolic pathway with additional metabolic engineering strategies, including the knockout of ADH2, downregulation of FPS1 and overexpression of the transcription regulatory gene Rim15. The fermentation characteristics of the engineered S. cerevisiae strain were investigated to analyze the effect of gene editing on the production of ethanol and by-products. Finally, we developed a novel genome-wide mutation system that accumulates distinct mutations in the industrial S. cerevisiae to improve robustness under stress conditions efficiently.
2. Materials and Methods
2.1. Strains, Medium and Culture Condition
E. coli DH5α was employed for plasmid construction and cultured in LB medium with 100 mg/L ampicillin. The industrial S. cerevisiae M (From Angel Yeast) was used as the parent strain routinely cultured in YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose). Recombinant S. cerevisiae strains were screened in YPD solid medium containing 50 mg/L nourseothricin, and 200 mg/L hygromycin B or 200 mg/L G418, and corn mash medium was used for the verification of yeast fermentation performance.
2.2. Plasmid Construction
The truncated fragments of the TEF promoter were cloned into the pSCM-N20 plasmids using Gibson assembly methods. The codon-optimized genes of ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) from Pseudomonas sp. and phosphoribulokinase (PRK) from spinach were synthesized by Jinkairui Biological Engineering Co., Ltd. (Wuhan, China). The codon-optimized Escherichia coli chaperones GroEL and GroES were synthesized by Jinkairui Biological Engineering Co., Ltd. (Wuhan, China). The key genes that regulate glycerol or ethanol yield, including FPS1 and the great wall-family protein kinases gene Rim15 were all amplified from the chromosome of the S. cerevisiae and subsequently cloned into the donor helper plasmids using digestion/ligation and/or Gibson assembly methods. Benchling CRISPR tool (https://benchling.com) was used to design gRNAs, which were cloned into Bsa I digested pSCM-gRNA. All the DNA polymerase, T4 DNA ligases, and restriction enzymes were purchased from New England Biolabs. Primers were synthesized by Tsingke Biotech Co., Ltd. (Wuhan, China).
All the plasmids used in this study were listed in Supplementary Table S1. All the primers used for plasmid construction and genome integration are listed in Supplementary Table S2. The coding sequences of heterologous genes are listed in Supplementary Table S3.
2.3. Strain Construction
The integration of related genes mentioned above into the chromosome of S. cerevisiae was performed through the CRISPR/Cas9 method [24]. The gene expression cassettes, together with the homology arms and the corresponding gRNA plasmids, were co-transformed into the S. cerevisiae strain harboring the SpCas9 plasmid. The ADH2 deletion was accomplished by substituting donor DNA containing 500 bp upstream and 500 bp downstream homologous arms of the target sequence, and the FPS1 promoter was replaced with the weak-intensity promoter MITp. The transformation of S. cerevisiae was performed using the LiAC method [25]. In this study, all the constructed strains were listed in Table 1.
2.4. Small-Scale Corn Fermentation
An engineered single colony strain was inoculated into YPD test tubes containing 5 mL of YPD medium and cultured overnight at 30 °C with a rotation speed of 200 r/min. Then all the cells in the test tube were transferred to a 500 mL shaker flask containing 200 mL YPD at the same culture conditions. After that, the cell culture is centrifuged at 5000 r/min for 5 min, and the supernatant is discarded. The cell pellet is resuspended by adding the sterile water equal to the mass.
The fermentation method was adopted from Khatibi et al. with some modifications [26]. Briefly, a corn mash consisting of 100 g of ground corn and 170 mL of water underwent a pretreatment process, in which 150 µL of amylase was added and stirred at 40 rpm for 90 min at a temperature of 101 °C. This pretreatment was intended to enhance the digestibility and subsequent utilization of the corn mash. Then 440 µL of glucoamylase and 640 µL of cell suspension were added to corn mash, and the net weight was fixed at 285 g by water. Fermentation flasks were incubated at 30 °C with 170 rpm for 72 h. At time points 0 h, 24 h, 48 h, and 72 h, the sample was weighed for analysis of weightlessness, and the flasks were immediately returned to the incubator. The alcohol content is determined at the end of fermentation at 72 h.
2.5. Analytical Methods
Once the fermentation process is finished, gently stir the fermentation mixture and then carefully pour 100 mL of it into a 1000 mL distillation flask. Simultaneously, 100 mL of tap water, along with two drops of defoamer, will be added to initiate the distillation process. The distilled liquid is then collected into a 100 mL volumetric flask using a cold water bath, ensuring that the temperature of the distillate remains below 25 ℃. A high-performance liquid chromatography (HPLC) system (Shimadzu, Kyoto, Japan) with a 300-mm Aminex HPX-87H column (0.6 mL/min, 5 mM H2SO4 at 65 °C; Bio-Rad, CA, USA) and a refractive index detector (Agilent, CA, USA) was used to detect fermentation metabolites.
2.6. Adaptive Evolution of Engineered Strains under the Multiple Stresses
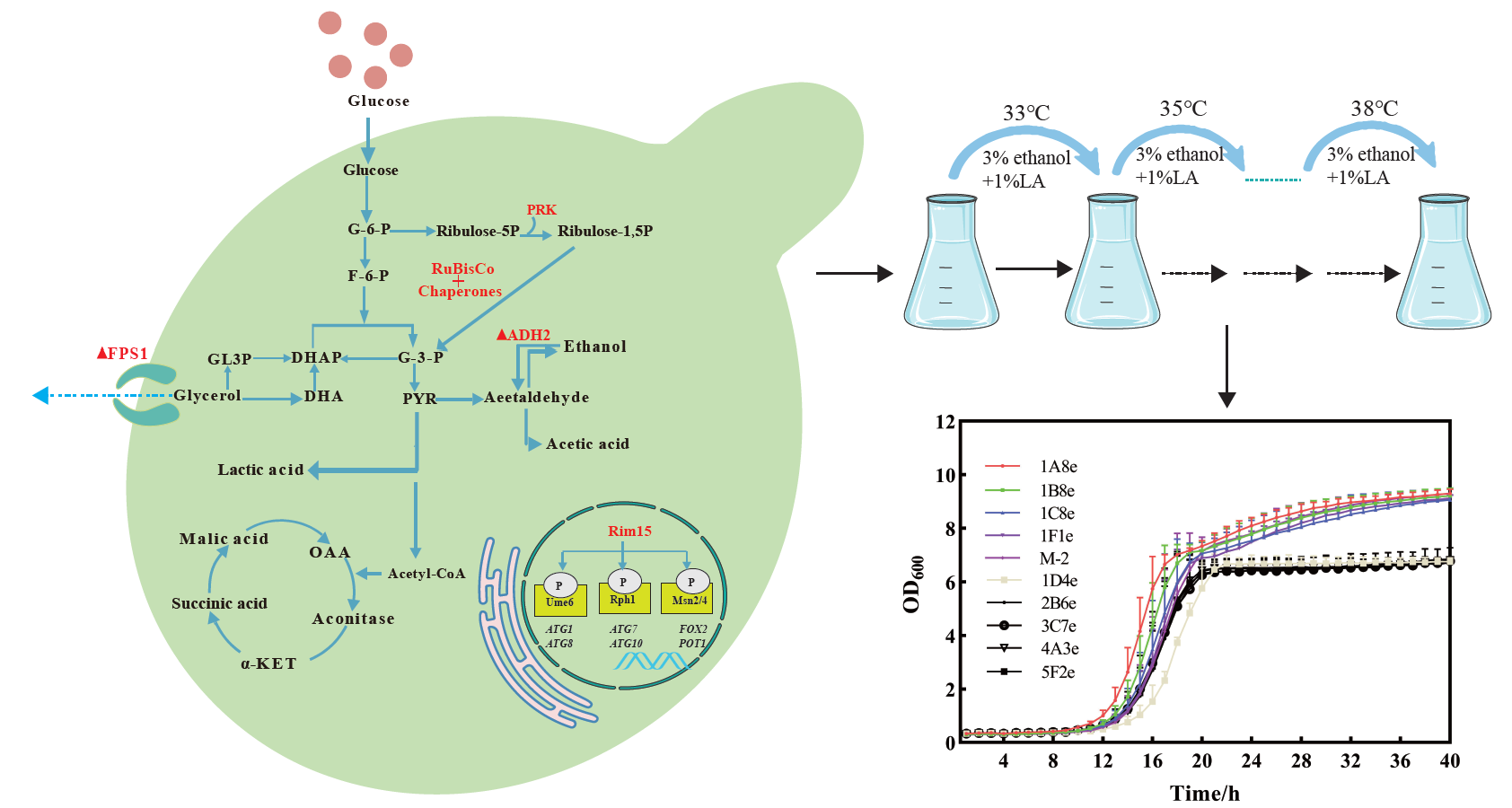
The adaptive evolution based on pH-dCas9-Mre11 plasmid system was introduced into the engineered S. cerevisiae by cultivating the cells at a temperature gradually increasing from 33 °C to 38 °C during 30 series of subculture, along with the addition of 1% lactic acid (LA) and 3% ethanol. The M-2 strain was grown for inoculum preparation until the late exponential phase was reached. This culture was used to inoculate flasks with YPD to an initial OD600nm of 0.2. When the culture reached the midlog phase, an aliquot was transferred to another flask with fresh medium. Each batch was started with a low initial biomass concentration (OD600nm ≈ 0.2) to select cells with better adaptability in 1% lactic acid and 3% ethanol, along with different temperatures.
Table 1. S. cerevisiae strains constructed in this study.
| Strain | Genotype | Source |
|---|---|---|
| M-1 | S. cerevisiae M(wild type) | From Angel yeast |
| M-2 | M-1-Site 21:: PGKp-PRK-ADH1t-CIT2p-Cbbm-CYC1t::Site 10::GAPp-GroES-CYC1t-TEF1p-GroEL-ADH1t | This study |
| M-3 | M-2-Site 5::CIT2p-Cbbm-CYC1t | This study |
| M-4 | M-1ΔADH2 | This study |
| M-5 | M-1-ΔFPSp::MIT1p | This study |
| M-6 | M-1-Site 19::TEF1p-Rim15-ADH1t | This study |
3. Results
3.1. Efficient Gene Editing in Industrial S. cerevisiae Using the CRISPR/Cas9 System via an Optimized gRNA Plasmid
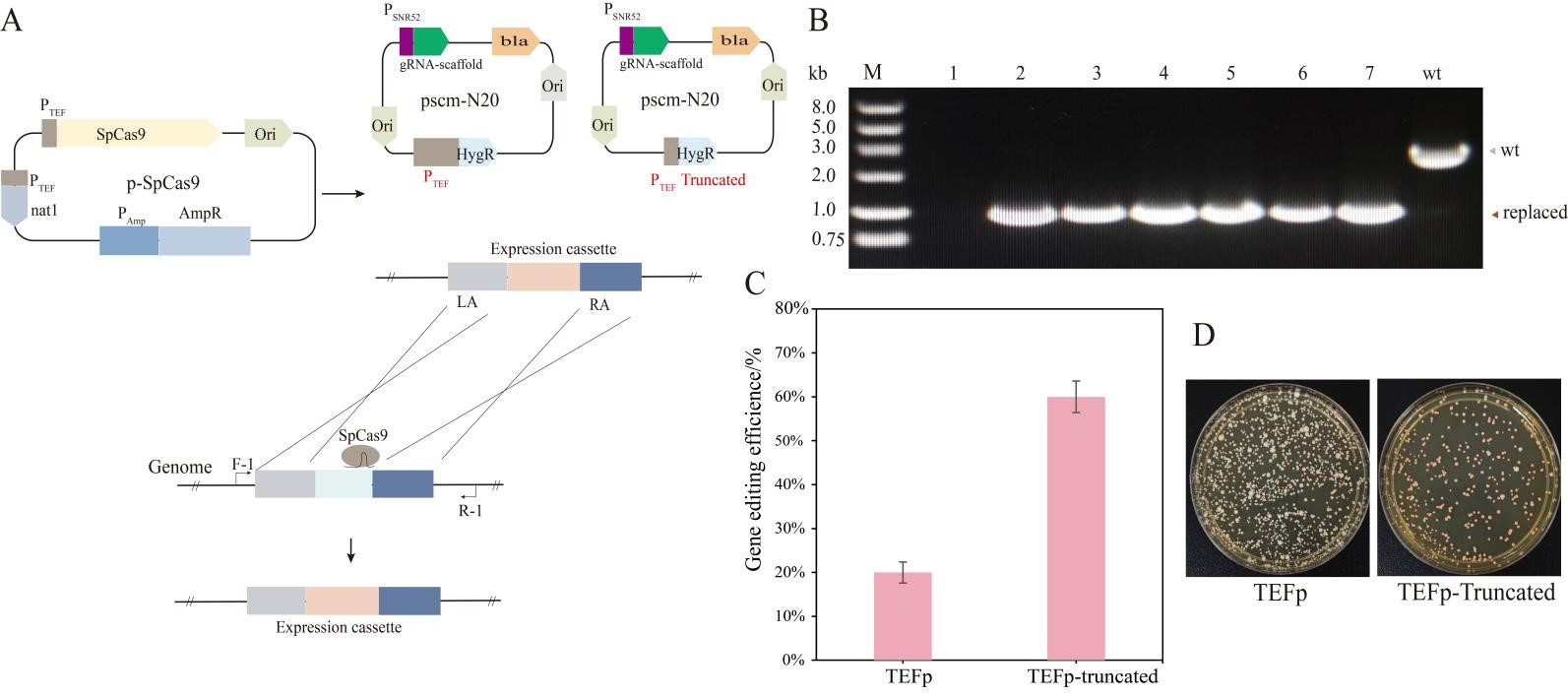
To improve the gene editing efficiency of industrial S. cerevisiae M, we used the CRISPR/Cas9 system via an optimized gRNA plasmid (Figure 1A,B). In this study, the copy number of optimized CRISPR-gRNA plasmid is regulated by a truncated TEF promoter. The ade2 gene was chosen to test the gene editing efficiency in industrial S. cerevisiae M by the optimized CRISPR system, of which the disruption of ade2 in S. cerevisiae resulted in a distinct pink colony phenotype caused by the accumulation of purine precursors in the vacuoles of yeast cells (Figure 1D). With the CRISPR-optimized gRNA plasmid system, the editing efficiency of the ade2 knockout in industrial S. cerevisiae increased to 60%, compared to only 20% in the control group (Figure 1C). The results showed that the optimized CRISPR/Cas9 system could significantly improve the gene editing efficiency of industrial S. cerevisiae M.
3.2. Multi-Strategies for the Construction of Engineered Yeast Strains for Reducing Glycerol Production
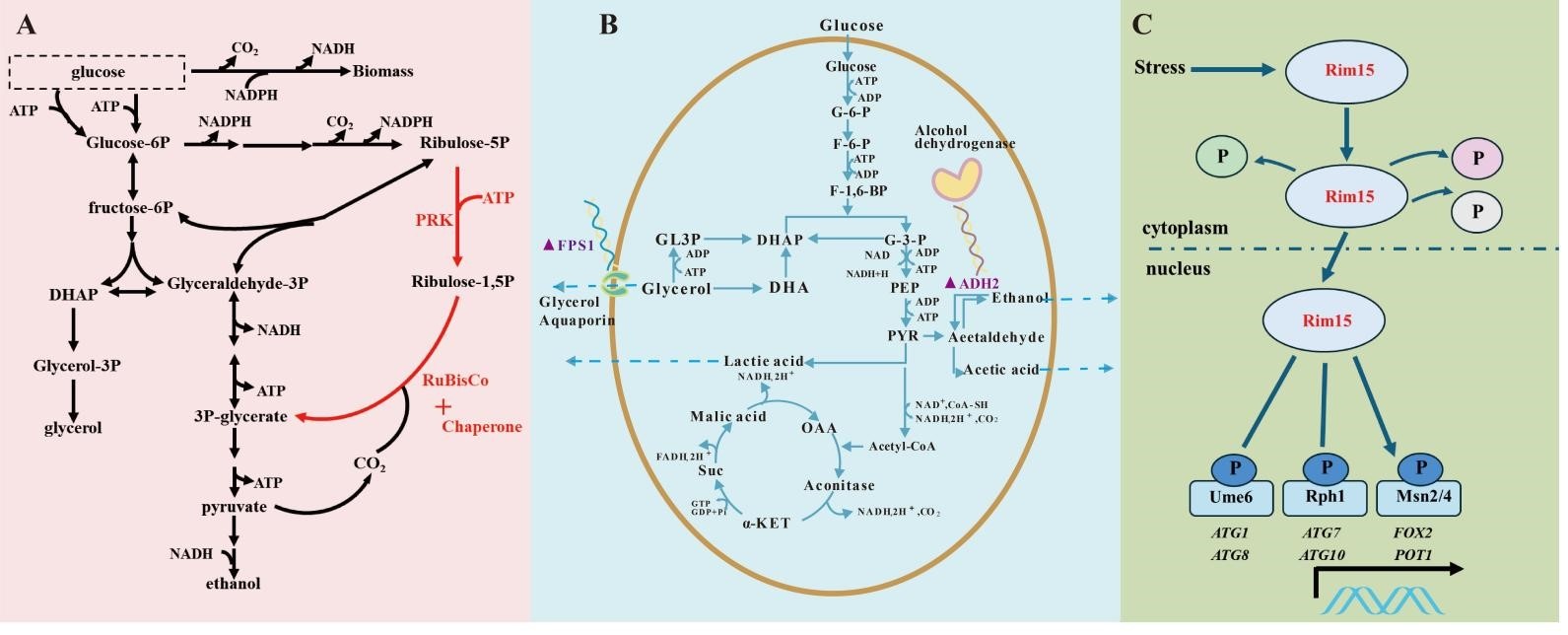
To reduce the production of glycerol, a CO2-fixation pathway was constructed in industrial S. cerevisiae M by heterologous expression of codon-optimized RuBisCO and PRK in combination with the codon-optimized chaperone GroEL/GroES from E. coli to obtain engineered strain M-2 (Figure 2A). Furthermore, the M-3 strain was obtained by increasing the two copies of RuBisCO gene to improve the efficiency of CO2 fixation further. In the S. cerevisiae, the enzyme known as alcohol dehydrogenaseII (ADH2) plays a crucial role in oxidizing ethanol to acetaldehyde. Therefore, decreasing the activity of ADH2 results in a lower consumption of ethanol during the engineering of metabolic pathways. Consequently, we successfully developed the engineered strain M-4 by deactivating the ADH2 gene (Figure 2B). Meanwhile, the S. cerevisiae ΔFPS1 mutant strain M-5 was constructed to achieve intracellular glycerol accumulation and trigger the reduction of glycerol biosynthesis in other regulatory systems, which then led to an increase in ethanol production (Figure 2B). Previous studies have shown the Rim15 gene as a crucial regulator in controlling the transcription levels of genes related to stress tolerance in yeast cells. To improve the yeast strains’ resistance to external environmental stress, we have successfully developed a new strain named M-6 through the overexpression of the Rim15 gene (Figure 2C).
3.3. Small-Scale Corn Fermentation for Lower Production of Glycerol
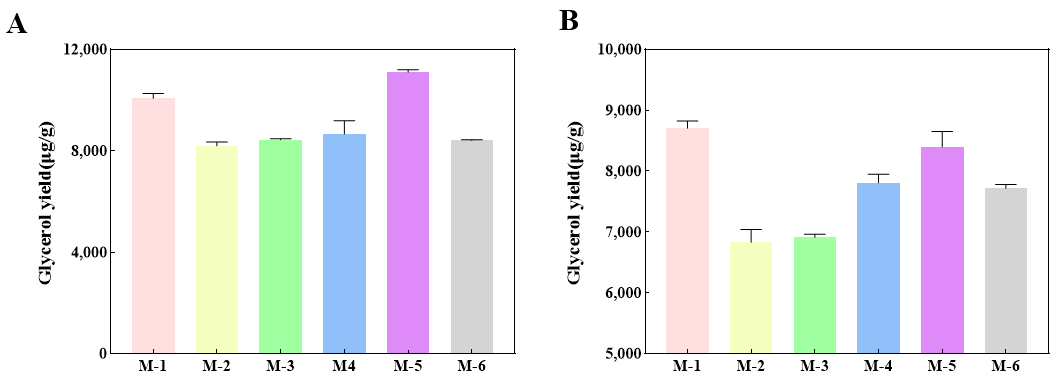
The yeast S. cerevisiae M used in this study is a commercial, industrial diploid yeast strain with relatively strong thermal tolerance, and the fermentation temperature can be maintained at up to 40 °C in practical production. Since the higher temperature is one of the key process parameters affecting fermentation efficiency, ethanol yield, and the formation of by-products (such as glycerol content). Therefore, we tested the fermentation parameters of the engineered yeast strains at 36 °C and 39 °C. The glycerol and ethanol yield in engineered strains mentioned above at the end of the fermentation were investigated compared to the wild-type strain. The results indicated that the glycerol content of the engineered strains decreased to some extent under fermentation conditions at 36 °C. Compared to the original strain, the glycerol content of M-2 significantly decreased by 18.6% (Figure 3A). Similarly, after raising the fermentation temperature to 39 °C, the glycerol yield of strains M-2 to M-6 underwent a reduction in varying magnitudes compared to the original strain. Among them, the glycerol content of M-2 significantly decreased by 21.5% in comparison to that of the original strain (Figure 3B).
However, the ethanol production of the engineered strain did not increase significantly compared with the original strain, and even the ethanol content of M-2, M-3, and M-6 decreased slightly at 36 ℃ and 39 ℃ (Figure S1B,D). Further analysis revealed that the cumulative weight loss of the engineered strains, except for M-4 was reduced in comparison to that of the original strain, indicating a decline in the growth performance of the strains (Figure S1A,C).
3.4. Construction and Characterization of Random Diversification in the S. cerevisiae Genome for Adaptive Evolution of Engineered Strains
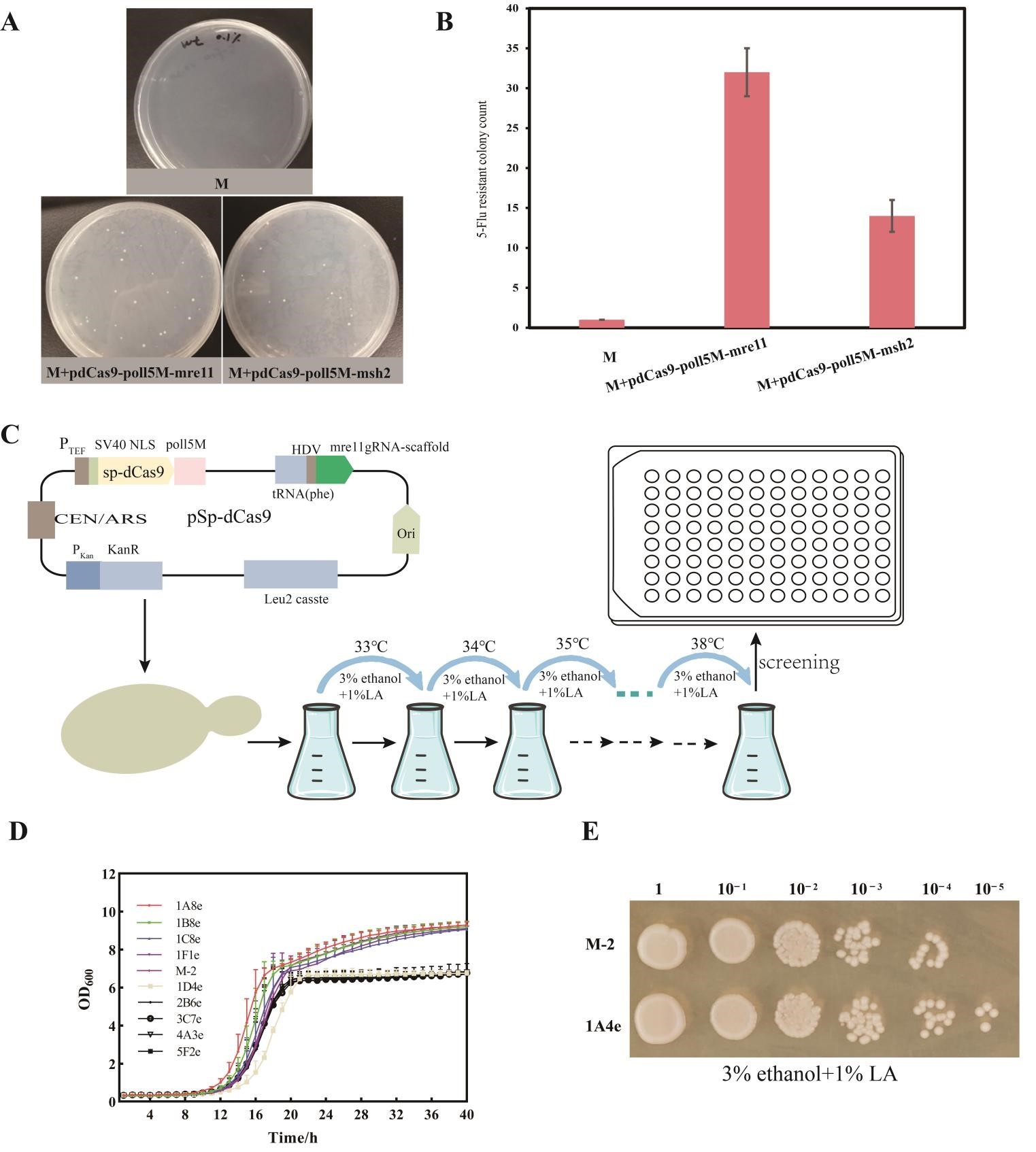
To further enhance the robustness of the engineered S. cerevisiae strains, genome maintenance genes mediated genome-wide mutations system (GWMS) serves as an effective method to diversify genes in their native context. In this study, the target gene upp locus, which encodes a uracil phosphoribosyltransferase whose inactivation ablates the ability of cells to uptake 5-fluorouracil (5-Flu), was tested. Only cells with upp disabled survive when grown on media containing 1.0 mg/mL 5-Flu, where the 5-Flu is not converted by a toxic 5-fluoro-2′-deoxyuridine-5′-monophosphate(5-F-dUMP). Thus, the upp gene serves as a negative selection to measure mre11-GWMS mutagenesis at this locus. Utilizing a gRNA targeting mre11 or msh2, liquid cultures were plated on SCD plates supplemented with 5-fluorouracil after 96 h of growth. 5-fluorouracil resistant (5-FluR) CFUs were quantified. The mre11 targeting mutation efficiency showed a 2.28-fold that of msh2 targeting mutation efficiency (Figure 4A,B). Next, we used mre11-GWMS for random diversification in the S. cerevisiae genome by adaptive evolution, which cultivating the cells at a temperature gradually increasing from 33 °C to 38 °C, along with the addition of 1% lactic acid (LA) and 3% ethanol (Figure 4C). After 30 series of subcultures, the cell growth of evolved strain 1A4e showed a significant increase compared with those of the unevolved strain (Figure 4D,E). However, there was no significant increase in ethanol production compared with the non-evolved strain (Figure S2).
In summary, these results indicate that Mre11-GWMS mediated adaptive evolution is an effective way to screen strains with good growth performance. However, further research is needed to improve the ethanol production of the strains.
Figure 1. Evaluation of the CRISPR/Cas9 editing efficiency with optimized gRNA plasmid. (<strong>A</strong>) Editing schematic of CRISPR-Cas system with different types of gRNA plasmids in industrial <em>S. cerevisiae</em>. (<strong>B</strong>) Disruption of the <em>ade2</em> gene mediated by the CRISPR/Cas9 system with a truncated TEF promoter of gRNA plasmid in industrial <em>S. cerevisiae </em>M. (<strong>C</strong>) Comparison of the editing efficiency with the truncated TEF promoter and conventional TEF promoter of gRNA plasmid. (<strong>D</strong>) Growth of strains with disruption of the <em>ade2</em> gene.
Figure 2. Metabolic engineering of industrial <em>S. cerevisiae</em> for reducing the glycerol content. Genes overexpressed and knocked-out in the present study were shown in red and purple, respectively. (<strong>A</strong>) Modification of the carbon metabolism pathway of <em>S. cerevisiae</em> is needed to reduce the ethanol yield in this study. (<strong>B</strong>) Schematic representation of central carbon metabolism and the introduced Calvin-cycle enzymes in <em>S. cerevisiae</em>. Phosphoribulokinase (PRK from <em>Spinacia oleracea</em>) and ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO form <em>Thiobacillus denitrificans</em>) in combination with the <em>E. coli</em> chaperones GroEL/GroES assist the folding of RuBisCO protein. (<strong>C</strong>) A schematic diagram showing the key genes related to Rim15-mediated regulation of stress response. In response to external stress, Rim15 is retained in the nucleus where it phosphorylates the transcriptional repressors (Ume6, Rph1) and activators (Msn2/4), promoting the upregulation of stress response genes.
Figure 3. The glycerol yield in engineered strains at the endpoint of the fermentation. (<strong>A</strong>) Glycerol yield in engineered strains fermented at 36 °C; (<strong>B</strong>) Glycerol yield in engineered strains fermented at 39 °C.
Figure 4. Enhancing the robustness of the engineered strain by genome maintenance gene <em>mre11 </em>mediated genome-wide mutations system adaptive evolution. (<strong>A</strong>) Gene mutators induced different numbers of colonies in the <em>S. cerevisiae </em>targeted the<em> upp </em>gene on the SCD medium containing 5-fluorouracil; (<strong>B</strong>) Gene mutators induced different relative mutation rates in the <em>S. cerevisiae </em>targeted the<em> upp </em>gene on the SCD medium containing 5-fluorouracil; (<strong>C</strong>) Schematic illustration of adaptive evolution and strain screening process; (<strong>D</strong>) The growth curve of different strains after evolution. (<strong>E</strong>) Ethanol and lactate tolerance after strain evolution.
4. Discussion
During the fermentation process for ethanol production, glycerol is typically formed as a by-product to maintain osmotic pressure and prevent water loss under high osmolarity conditions [27]. However, excessive accumulation will inevitably have a detrimental effect on ethanol production [27]. To date, many researchers have utilized CRISPR-Cas9 technology to construct engineered strains of S. cerevisiae for the regulation of glycerol production [28,29]. Functional expression of the PRK and Rubisco in S. cerevisiae led to a 90% reduction of the by-product glycerol and a 10% increase in ethanol production in sugar-limited chemostat cultures on a mixture of glucose and galactose [30]. The deletion of FPS1 increased ethanol production by 10% and reduced glycerol yield by 18.8% [14], and ADH2 deletion in S. cerevisiae also resulted in the improvement of ethanol yield [28]. In this study, the glycerol production of engineered strains M-2, M-3, M-4 and M-6 decreased to different degrees, in which the glycerol production of M-2 decreased by 21.5% (Figure 3), but the ethanol production did not exhibit a significant increase. Further analysis of the strains’ cumulative weight loss revealed a decrease in all cases, likely due to reduced growth performance, which hindered ethanol production.
Adaptive evolution has been proven to be an effective method for selecting yeast populations resistant to various stress conditions and expanding their tolerance range [30,31]. With the development of new strategies and tools for adaptive evolution, the efficiency of modifying metabolic pathways and enhancing cellular tolerance has been significantly accelerated [32]. To further enhance the growth performance of the strain, the adaptive evolution based on the mre11-GWMS was performed. The cell growth of evolved strains showed better growth performance compared with those of the unevolved strain (Figure 4D), indicating that the adaptive evolution based on the mre11-GWMS is an effective method to improve the tolerance and growth ability of strains. However, with the progression of adaptive evolution technologies, the process of constructing and screening cell factories frequently encounters limitations imposed by the assessment of targeted phenotypes. The greatest challenge consists of developing high-throughput screening methods to identify the optimal mutants from millions of potential candidates. Further improvements and optimizations of existing technologies are necessary to convert difficult-to-detect phenotypes into rapidly measurable ones, integrating adaptive evolution with high-throughput screening to drive the further development of this technology.
In this study, we optimized the gene editing efficiency of industrial S. cerevisiae and reduced the glycerol content of engineered strains to varying degrees through various metabolic engineering strategies. The resulting CO2-fixing yeast M-2 led to a 21.5% reduction of the by-product glycerol in corn mash fermentation cultures at 39 ℃, and further adaptive evolution based on the mre11-GWMS can effectively improve the strain’s tolerance and growth performance.
Supplementary Materials
The following supporting information can be found at: https://www.sciepublish.com/article/pii/462, Figure S1. Cumulative weightlessness and ethanol yield in engineered strains. (A) Difference in cumulative weightlessness at 36 ℃. (B) Difference in ethanol yield at 36 ℃. (C) Difference in cumulative weightlessness at 39 ℃. (D) Difference in ethanol yield at 39 ℃; Figure S2. The ethanol yield of the evolved strains and the parental strain. (A) Ethanol yield of the evolved strains and the parental strain fermented at 35 °C. (B) Ethanol yield of the evolved strains and the parental strain fermented at 38 °C; Figure S3. Residual sugar and acetic acid in fermentation of engineered strains. (A) Residual sugar and acetic acid in fermentation of engineered strains at 36 °C. (B) Residual sugar and acetic acid in fermentation of engineered strains at 39 ℃; Table S1. A list of plasmids constructed in this study; Table S2. A list of primers used in this study; Table S3. A list of heterologous genes coding sequences used in this study.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
Author Contributions
N.X. and X.G. designed the study, analyzed the data, and wrote the paper. N.X., Y.Y., H.C., Y.Z., B.L., N.P. and Y.W. conducted the experiments. All the authors approved the manuscript.
Ethics Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Funding
This work was financially supported by the Hubei Provincial Natural Science Foundation Yichang Innovation and Development Joint Fund Project (2024AFD125).
Declaration of Competing Interest
The authors declare no competing financial interest.
References
1.
Nissen TL, Hamann CW, Kielland-Brandt MC, Nielsen J, Villadsen J. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast 2000, 16, 463–474. [Google Scholar]
2.
Guo Z-P, Zhang L, Ding Z-Y, Shi G-Y. Minimization of glycerol synthesis in industrial ethanol yeast without influencing its fermentation performance. Metab. Eng. 2011, 13, 49–59. [Google Scholar]
3.
Nissen TL, Kielland-Brandt MC, Nielsen J, Villadsen J. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab. Eng. Commun. 2000, 2, 69–77. [Google Scholar]
4.
Bro C, Regenberg B, Forster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 2006, 8, 102–111. [Google Scholar]
5.
Van Dijken JP, Scheffers WA. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol. Rev. 1986, 32, 199–224. [Google Scholar]
6.
Guadalupe-Medina V, Wisselink HW, Luttik MA, de Hulster E, Daran JM, Pronk JT, et al. Carbon dioxide fixation by Calvin Cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels 2013, 6, 125. [Google Scholar]
7.
Liu Z, Wang K, Chen Y, Tan T, Nielsen J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 2020, 3, 274–288. [Google Scholar]
8.
Santos Correa S, Schultz J, Lauersen KJ, Soares Rosado A. Natural carbon fixation and advances in synthetic engineering for redesigning and creating new fixation pathways. J. Adv. Res. 2023, 47, 75–92. [Google Scholar]
9.
Li Y-J, Wang M-M, Chen Y-W, Wang M, Fan L-H, Tan T-W. Engineered yeast with a CO2-fixation pathway to improve the bio-ethanol production from xylose-mixed sugars. Sci. Rep. 2017, 7, 43875. [Google Scholar]
10.
Xia P-F, Zhang G-C, Walker B, Seo S-O, Kwak S, Liu J-J, et al. Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth. Biol. 2016, 6, 276–283. [Google Scholar]
11.
Papapetridis I, Goudriaan M, Vázquez Vitali M, de Keijzer NA, van den Broek M, van Maris AJA, et al. Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol. Biofuels 2018, 11, 17. [Google Scholar]
12.
Yang P, Jiang S, Lu S, Jiang S, Jiang S, Deng Y, et al. Ethanol yield improvement in Saccharomyces cerevisiae GPD2 Delta FPS1 Delta ADH2 Delta DLD3 Delta mutant and molecular mechanism exploration based on the metabolic flux and transcriptomics approaches. Microb. Cell Factories 2022, 21, 160. [Google Scholar]
13.
Tamás MJ, Luyten K, Sutherland FCW, Hernandez A, Albertyn J, Valadi H, et al. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 2002, 31, 1087–1104. [Google Scholar]
14.
Zhang A, Kong Q, Cao L, Chen X. Effect of FPS1 deletion on the fermentation properties of Saccharomyces cerevisiae. Lett. Appl. Microbiol. 2007, 44, 212–217. [Google Scholar]
15.
Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 2011, 477, 471–476. [Google Scholar]
16.
DiCarlo JE, Conley AJ, Penttilä M, Jäntti J, Wang HH, Church GM. Yeast Oligo-Mediated Genome Engineering (YOGE). ACS Synth. Biol. 2013, 2, 741–749. [Google Scholar]
17.
Jones S. MAGE in yeast is a go. Nat. Biotechnol. 2017, 35, 1147–1148. [Google Scholar]
18.
Bao Z, HamediRad M, Xue P, Xiao H, Tasan I, Chao R, et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision. Nat. Biotechnol. 2018, 36, 505–508. [Google Scholar]
19.
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, 6305. [Google Scholar]
20.
Tou CJ, Schaffer DV, Dueber JE. Targeted diversification in the S. cerevisiae genome with CRISPR-guided DNA polymerase I. ACS Synth. Biol. 2020, 9, 1911–1916. [Google Scholar]
21.
Le Borgne S. Genetic engineering of industrial strains of Saccharomyces cerevisiae. Recomb. Gene Expr. 2012, 824, 451–465. [Google Scholar]
22.
Stovicek V, Borodina I, Forster J. CRISPR–Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab. Eng. Commun. 2015, 2, 13–22. [Google Scholar]
23.
Lian J, Bao Z, Hu S, Zhao H. Engineered CRISPR/Cas9 system for multiplex genome engineering of polyploid industrial yeast strains. Biotechnol. Bioeng. 2018, 115, 1630–1635. [Google Scholar]
24.
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar]
25.
Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006, 313, 107–120. [Google Scholar]
26.
Khatibi PA, Roach DR, Donovan DM, Hughes SR, Bischoff KM. Saccharomyces cerevisiae expressing bacteriophage endolysins reduce Lactobacillus contamination during fermentation. Biotechnol. Biofuels 2014, 7, 104. [Google Scholar]
27.
Nevoigt E, Stahl U. Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 1997, 21, 231–241. [Google Scholar]
28.
Xue T, Liu K, Chen D, Yuan X, Fang J, Yan H, et al. Improved bioethanol production using CRISPR/Cas9 to disrupt the ADH2 gene in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2018, 34, 154. [Google Scholar]
29.
Yang P, Jiang S, Jiang S, Lu S, Zheng Z, Chen J, et al. CRISPR-Cas9 approach constructed engineered Saccharomyces cerevisiae with the deletion of GPD2, FPS1, and ADH2 to enhance the production of ethanol. J. Fungi 2022, 8, 703. [Google Scholar]
30.
Sha Y, Zhou L, Wang Z, Ding Y, Lu M, Xu Z, et al. Adaptive laboratory evolution boosts Yarrowia lipolytica tolerance to vanillic acid. J. Biotechnol. 2023, 367, 42–52. [Google Scholar]
31.
Randez-Gil F, Prieto JA, Rodríguez-Puchades A, Casas J, Sentandreu V, Estruch F. Myriocin-induced adaptive laboratory evolution of an industrial strain of Saccharomyces cerevisiae reveals its potential to remodel lipid composition and heat tolerance. Microb. Biotechnol. 2020, 13, 1066–1081. [Google Scholar]
32.
Cao M, Tran VG, Zhao H. Unlocking nature’s biosynthetic potential by directed genome evolution. Curr. Opin. Biotechnol. 2020, 66, 95–104. [Google Scholar]